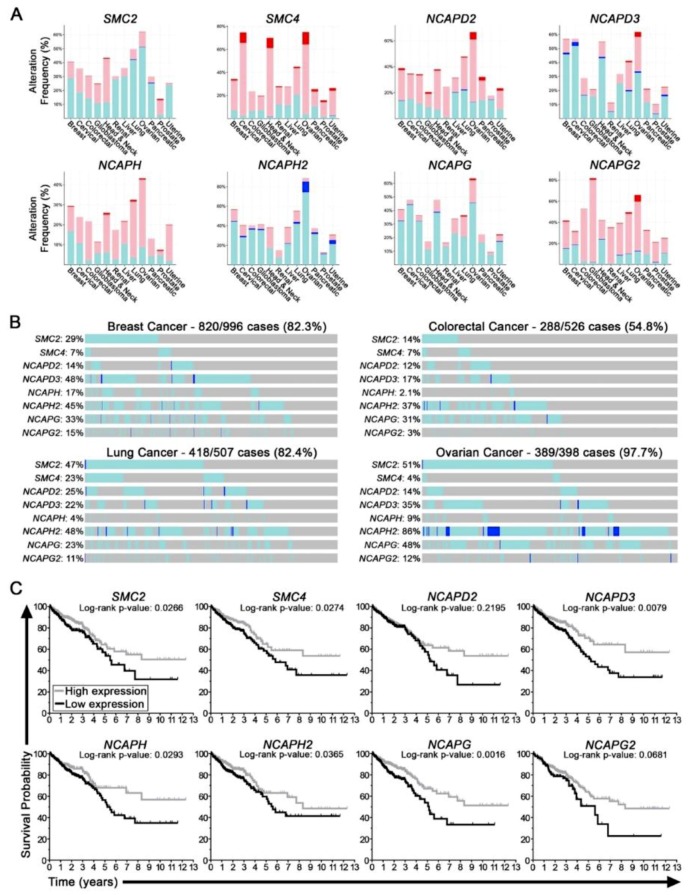
Figure 1.
Condensin gene alterations correspond with reduced expression and worse patient survival in cancer. (A) Frequency of condensin gene copy number alterations in 12 common cancer types [23]; shallow deletions (aqua), deep deletions (blue), small-scale gains (pink) and large-scale amplifications (red). (B) The cumulative frequency of shallow and deep deletions for all condensin genes in breast (82.3%), colorectal (54.8%), lung (82.4%), and ovarian (97.7%) cancers [23]. (C) Kaplan–Meier curves reveal that CRCs with reduced condensin gene expression (mRNA) typically correlate with worse overall survival relative to those with high expression [23]. Log-rank tests identify significantly worse outcomes for all genes, with the exception of NCAPD2 (p = 0.2195) and NCAPG2 (p = 0.0681), which do show similar trends.