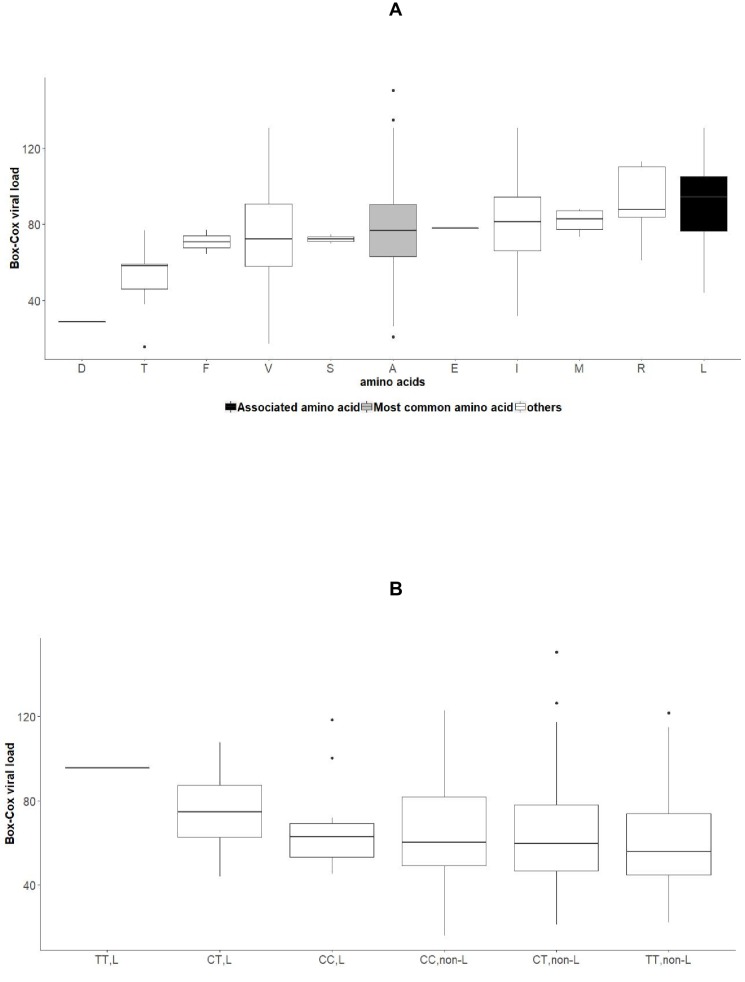
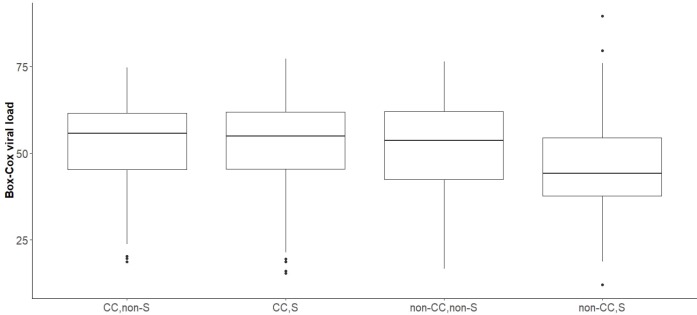
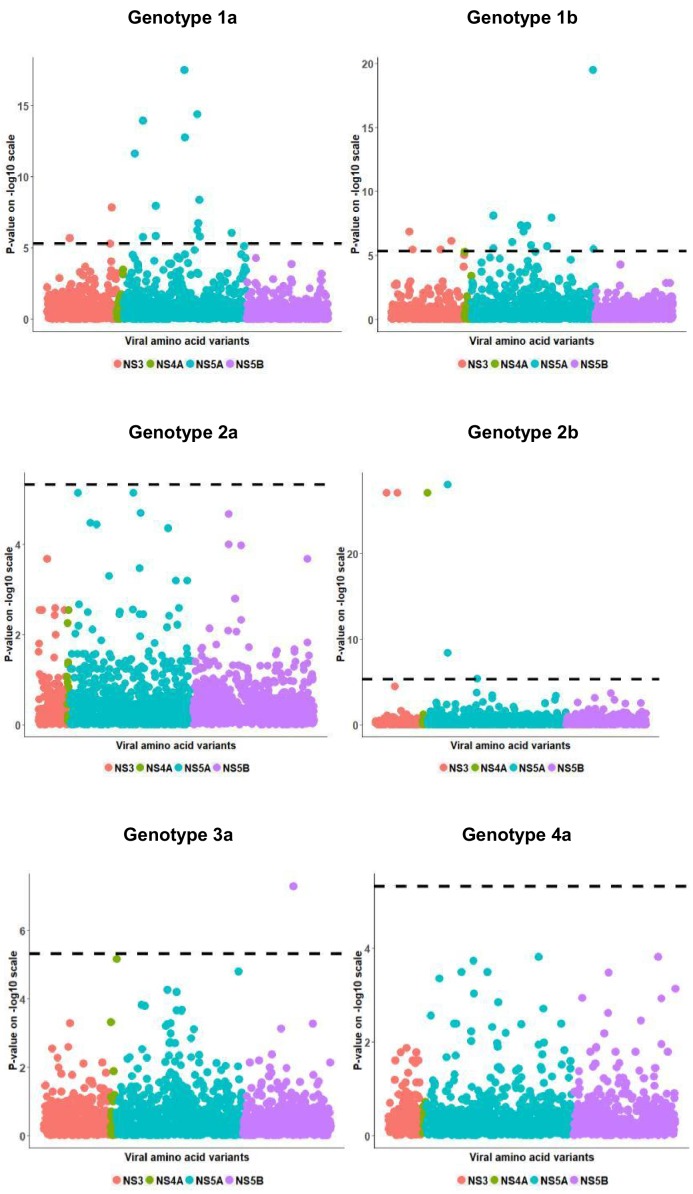
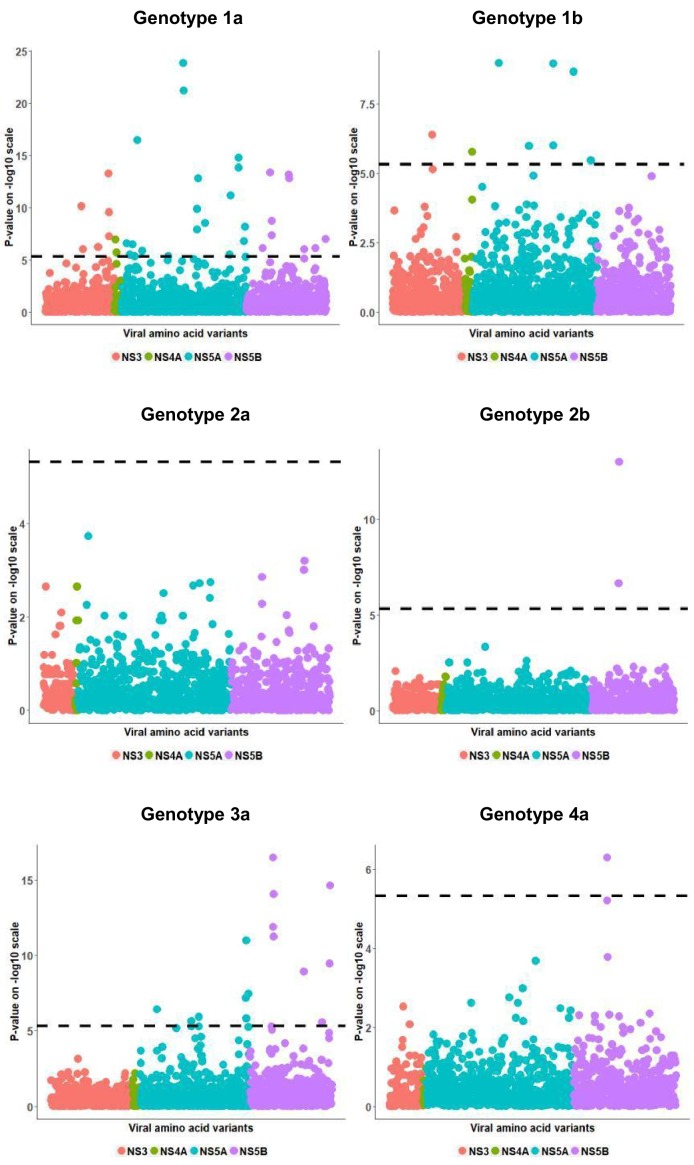
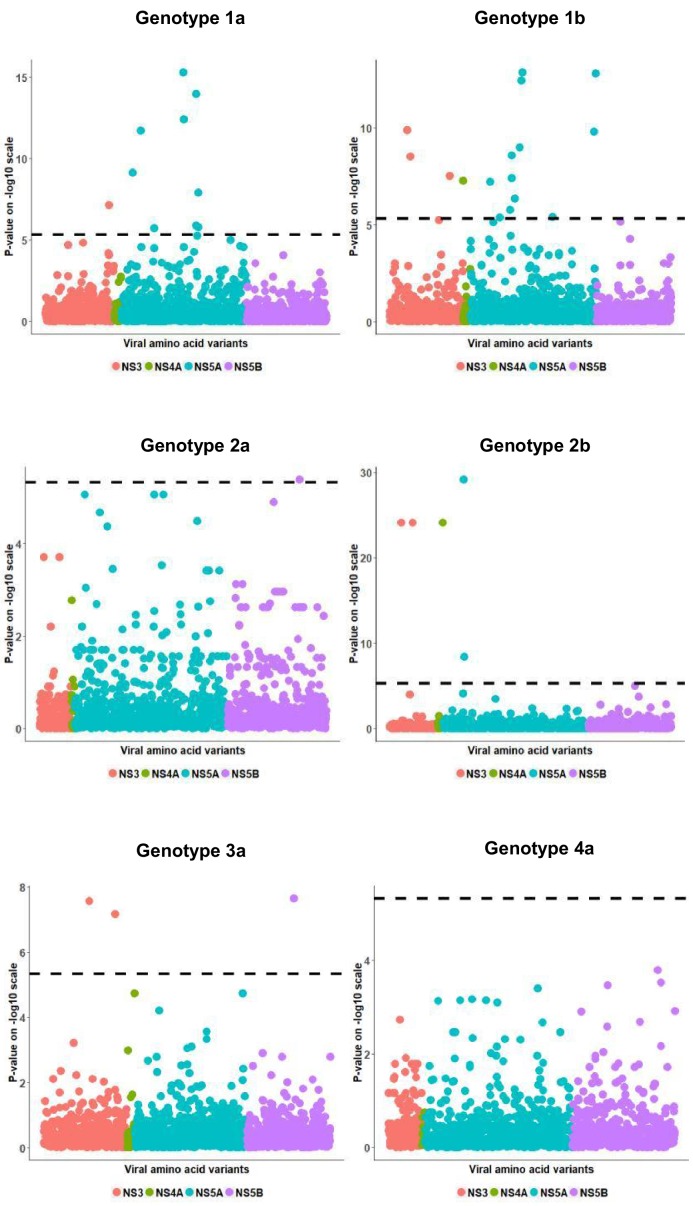
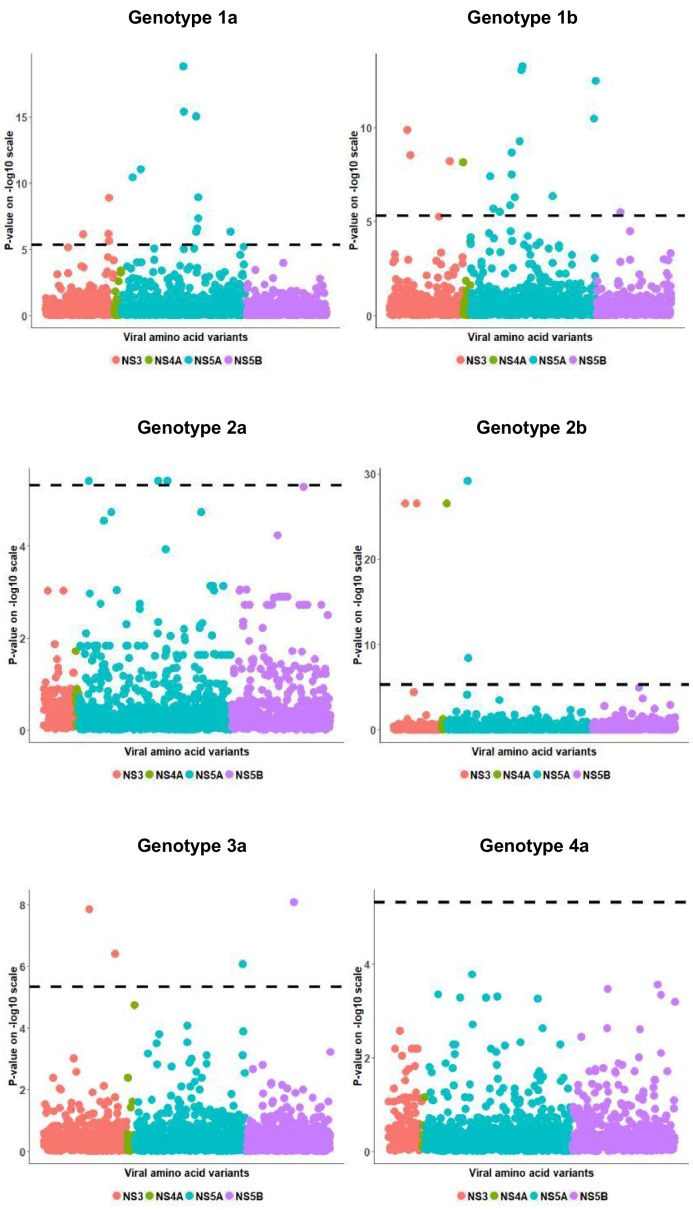
Figure 3. Associations between amino acid variables at position 2224 of NS5A, rs12979860 genotypes and HCV viral load in the group of patients infected with HCV genotype 1b.
(A) Boxplot of transformed viral load stratified by amino acids present at position 2224 of NS5A. (B): Boxplot of transformed viral load stratified by rs12979860 genotypes (CC, CT, TT) and by presence or absence of leucine at position 2224 of NS5A.