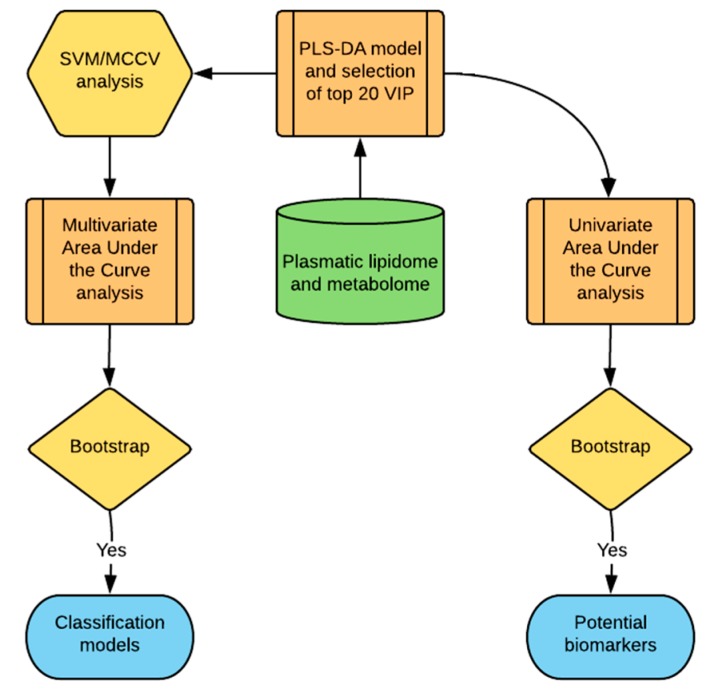
Figure 1.
Statistical approach used for the identification of potential lipid and metabolite-based biomarkers for the prediction of the alloreactivity following stem cell transplantation (SCT). A supervised statistical approach in the form of partial least squares discriminant analysis (PLS-DA) was applied to the consolidated metabolomic and lipidomic data as a first step to find potential biomarkers and to detect the presence of class separation, if any, between graft vs. host disease (GVHD) and non-GVHD patients. The top 20 most important variables, which separates the groups, were selected by the variable importance in projection (VIP). A support vector machine (SVM) used these VIP variables to further find the binary classification of patients in the two groups. The result was cross-validated with Monte Carlo cross-validation (MCCV) and was used in a multivariate area under the curve of the receiver operating characteristic (AUCROC) analysis to select the best model based on low dimensionality and high accuracy. Model estimates were validated with bootstrap CI. The best model was used as reference to select potential biomarkers of GVHD through univariate analysis of the top 20 VIP that passed the criteria of high AUCROC estimate and low t-test p-value, and validated with bootstrap CI.