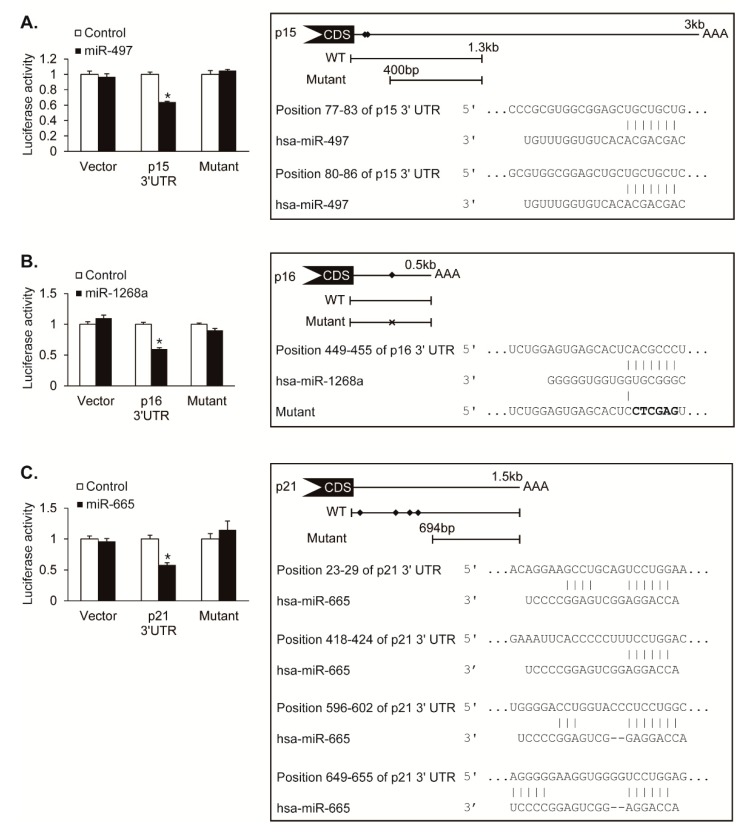
Figure 4.
Hypoxia-induced miRNAs target CDK inhibitors. (A–C) (Left panel) Luciferase activity of constructs with the 3′UTR of p15, p16, or p21 including the wild-type miRNA MREs, the mutant 3′UTR deleted miRNA recognition element (MRE) for miR-497 or miR-665, or the mutant 3′UTR with mutated MRE sequence for miR-1268a were examined in Cos7 cells by transfecting control, miR-497, miR-1268a, or miR-665 mimic. A luciferase vector without 3′UTR sequence (Vector) was used as a negative control. Data represent the mean ± S.E. of triplicates. * p < 0.05. (Right panel) Schematic diagrams of the predicted MREs of miR-497, miR-1268a, and miR-665 in the 3′UTR of p15, p16 and p21 transcripts, and the luciferase reporter constructs used for luciferase assays. The predicted MREs are marked with diamonds. CDS and AAA stand for protein coding sequence and poly(A) tail, respectively. Mutations introduced in the MRE to disrupt a base pairing with miR-1268a sequence are indicated as X. Sequences of the 3′UTR of CDK inhibitors and the predicted MRE of miRNAs are shown. Perfect base matches are indicated by a line.