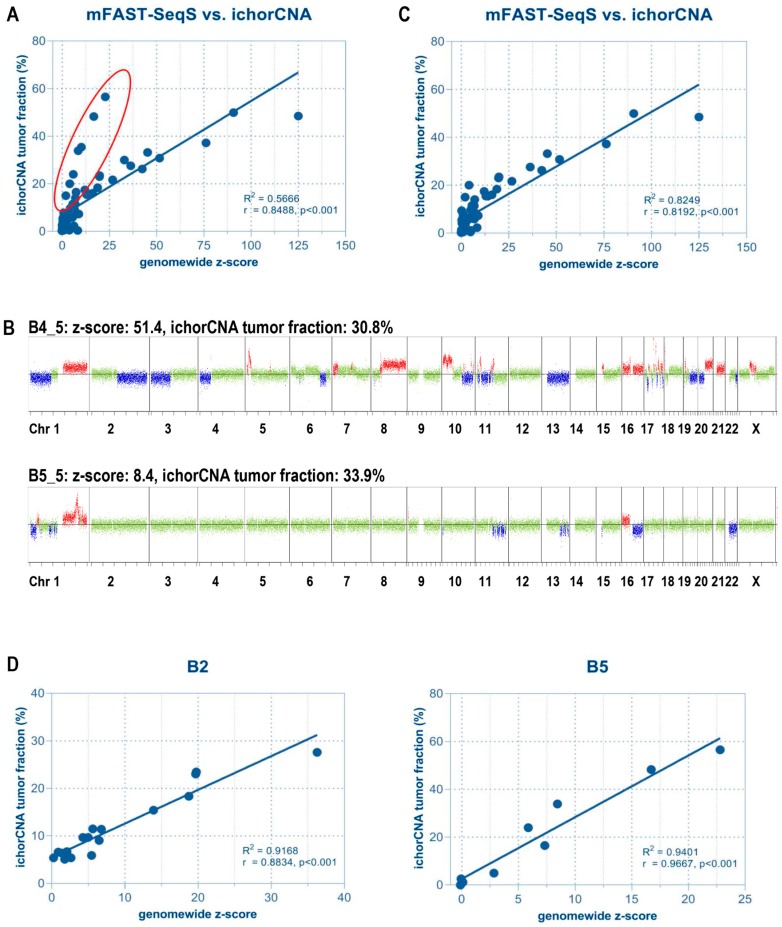
Figure 1.
Correlation between mFast-SeqS based z-scores and ichorCNA tumor fractions. (A) With the exception of some samples (highlighted in red), the majority showed a high correlation between z-score and tumor fraction. Most of these outliers were obtained from patient B5. (B) Samples with high tumor fraction but lower z-scores revealed a lower number of copy number alterations (CNAs) compared to other samples as exemplified by two genome-wide copy number profiles. Based on ichorCNA, both samples had a tumor fraction of approximately 30%. However, due to the low abundance of CNAs in patient B5, the mFAST-SeqS z-score was only 8.4, compared to a value of 51.4 of a patient with many CNAs across the genome. Red indicates a gain of chromosomal material, blue indicates a loss of chromosomal material, green depicts balanced regions. (C) The removal of samples with low overall amounts of CNA significantly improved the correlation. (D) Correlation between mFAST-SeqS z-scores and ichorCNA-based tumor fractions in selected individual patients.