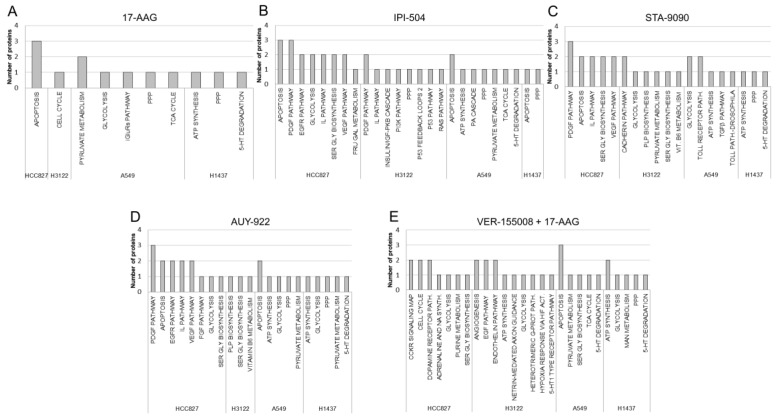
Figure 3.
Canonical pathway analysis of proteins identified by two-dimensional electrophoresis and mass spectrometry (2-DE/MS) in response to HSP90 inhibition. Functional analysis of the differentially expressed proteins in response to (A) 17-AAG, (B) IPI-504, (C) STA-9090, (D) AUY-922 and (E) VER-155008 + 17-AAG inhibitors in the adenocarcinoma cell lines HCC827, H3122, A549 and H1437. Enriched pathways were determined from the PANTHER database. The number of proteins involved in each identified pathway is represented on the y axis. iGluRs pathway = ionotropic glutamate receptor pathway, PPP= pentose phosphate pathway, TCA cycle = tricarboxylic acid cycle, 5-HT degradation = 5-hydroxytryptamine or serotonin degradation, PDGF pathway = platelet-derived growth factor signaling pathway, EGFR pathway = epidermal growth factor receptor pathway, IL pathway = interleukin signaling pathway, SER GLY biosynthesis = serine and glycine biosynthesis, VEGF pathway = vascular endothelial growth factor signaling pathway, FRU GAL metabolism = fructose and galactose metabolism, insulin/IGF-PKB cascade = insulin/ insulin-like growth factor pathway-protein kinase B signaling cascade, PI3K pathway = phosphoinositide-3 kinase pathway, PA cascade = plasminogen activating cascade, PLP biosynthesis = pyridoxal-5-phosphate biosynthesis, TGFβ pathway = transforming growth factor beta signaling pathway, FGF pathway = Fibroblast Growth Factor signaling pathway, adrenaline and NA synth. = adrenaline and noradrenaline biosynthesis, EGF pathway = epidermal growth factor pathway, heterotrimeric G-protein signaling pathway = heterotrimeric G-protein signalling pathway Gi alpha- and Gs alpha-mediated pathway, 5-HT1 type receptor pathway = serotonin 1A receptor-mediated signalling pathway, MAN metabolism = mannose metabolism.