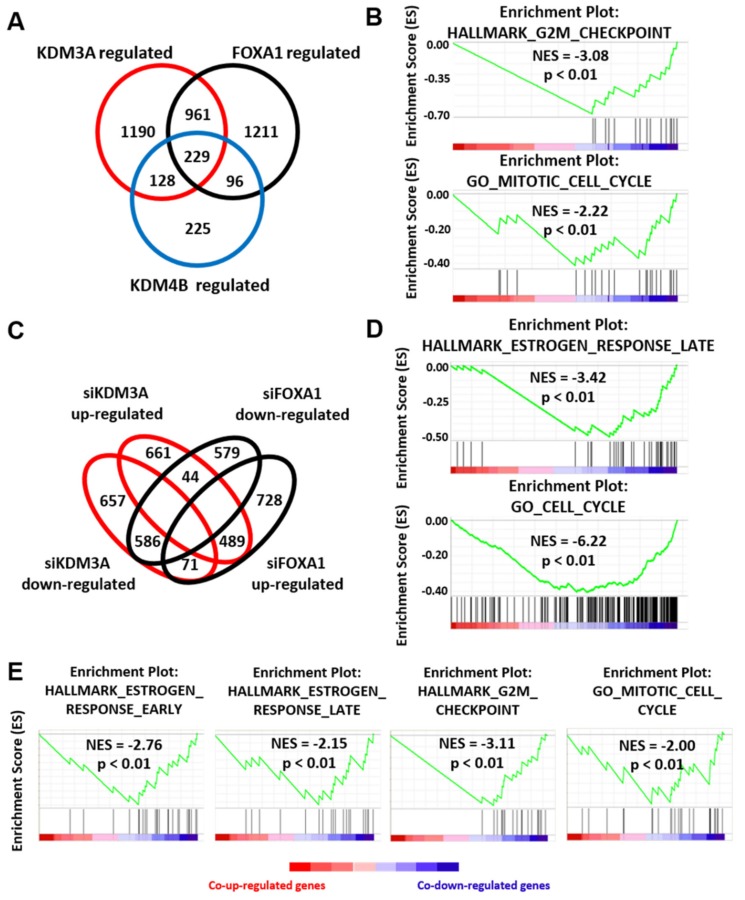
Figure 3.
KDM3A, KDM4B, and FOXA1 have overlapping gene regulatory profiles. (A) MCF-7 cells were transiently transfected with either a non-silencing control siRNA (siSCR), siKDM3A#1, siKDM4B#1, or siFOXA1#1 and grown in steroid-depleted medium for 48 h prior to treatment with 10 nM E2. RNA was extracted and samples hybridized to an Illumina HT12.2 Bead CHIP array. The number of KDM3A- (n = 2508), KDM4B- (n = 678), and FOXA1-regulated (n = 2497) genes (defined as genes significantly (p < 0.05) up or down regulated 1.5 fold upon knockdown compared to siSCR controls) was determined. Genes up- or down-regulated by more than one enzyme were classified as KDM3A/KDM4B/FOXA1 co-regulated genes (n = 1190 KDM3A/FOXA1 co-regulated genes; n = 357 KDM3A/KDM4B co-regulated genes; 325 KDM4B/FOXA1 co-regulated genes; n = 229 KDM3A/KDM4B/FOXA1 co-regulated genes). (B) GSEA of KDM3A/KDM4B/FOXA1 co-regulated genes demonstrates significant negative enrichment of genes associated with the HALLMARK_G2M_CHECKPOINT geneset and GO_MITOTIC_CELL_CYCLE geneset. NES = Nomalised Enrichment Score. p = nominal p-value. (C) Comparison between KDM3A- and FOXA1- up- and down-regulated genesets. (D) GSEA of KDM3A/FOXA1 co-regulated genes demonstrates significant negative enrichment of genes associated with the HALLMARK_ESTROGEN_RESPONSE_LATE geneset and GO_CELL_CYCLE geneset. (E) GSEA of KDM3A/KDM4B co-regulated genes demonstrates significant negative enrichment of genes associated with the HALLMARK_ESTROGEN_RESPONSE_EARLY, HALLMARK_ESTROGEN_RESPONSE_LATE and HALLMARK_G2M_CHECKPOINT genesets and the GO_MITOTIC_CELL_CYCLE geneset.