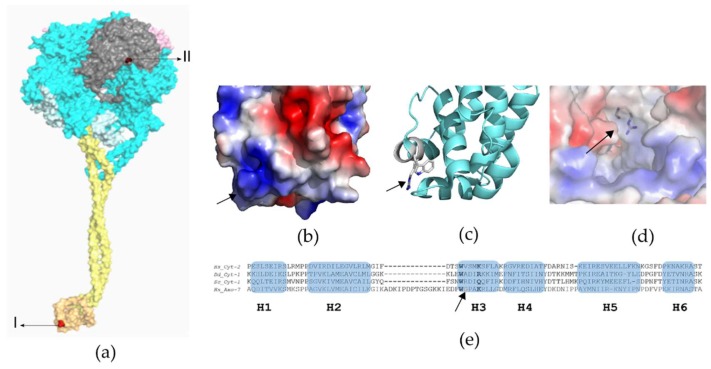
Figure 3.
Structural analysis of DNAH7 variants. (a) General dynein motor domain structure: N-terminal linker (pink), six AAA domains in ring arrangement (blue), stalk (yellow) containing the MTBS (orange), C-terminal (grey). Mutations (red) p.Gly2737Ser (I) and p.Arg3983trp (II). Crystallographic model PDB ID 4RH7: Crystallographic structure of human cytoplasmic dynein 2 motor domain. (b,c) Crystal structure of the MTBS of D. discoideum cytoplasmic dynein-1 (PDB ID: 3VKH): The p.Gly2737Ser mutation is in H3 (arrows). H3 is highly polar and assumes an important role in the interaction with the microtubule: electrostatic surface shown in (b) and H3 key residues in (c). (d) Close-view of the C-terminal of the crystallographic structure of human cytoplasmic dynein-2 motor domain (PDB ID 4RH7). The R side chain is disordered in the 4RH7 model as it is exposed to solvent. The putative position (arrow) of the p.Arg3983trp mutation highlights the superficial position of the residue. (e) Multiple sequence alignments: Hs_Cyt-2-human cytoplasmic dynein-2 (PDB ID: 4RH7), Dd_Cyt-1-D. discoideum cytoplasmic dynein-1 (PDB ID: 3VKG), Sc_Cyt-1-S. cerevisiae cytoplasmic dynein-1 (PDB ID: 4AKI), and Hs_Axo-7- human axonemal dynein-7. The secondary structure of Hs_Cyt-2 is indicated (alpha-helices H1 to H6). The arrow is pointing to the amino acid (glycine, G) that is mutated in Patient-2.