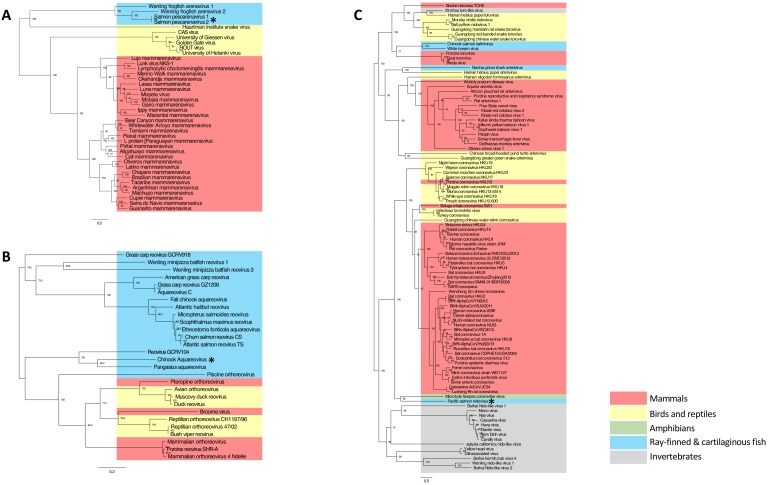
Figure 1. Maximum likelihood phylogenetic tree based on MAFFT alignments of the predicted replicase protein of (A) Salmon pescarenavirus and related arenaviruses, (B) Chinook Aquareovirus and related Aqua and orthoreoviruses and (C) Pacific salmon nidovirus and related Nidovirales.
Sequences from this study are marked with an asterisk, scale bar represents the number of amino substitutions per site, node labels show the bootstrap support and host groups are shown by colour. Trees are mid-point rooted, so do not necessarily represent the ancestral relationship of the viruses. Amino acid alignments have been provided in the source data for Figure 1.