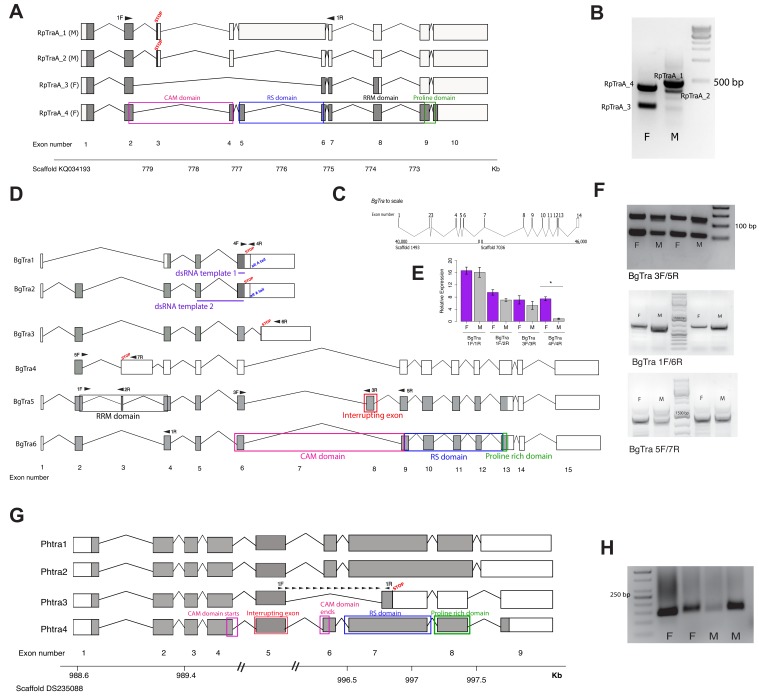
Figure 1. Transformer splicing follows the holometabolous pattern in the kissing bug Rhodnius prolixus, but not in the cockroach Blattella germanica or louse Pediculus humanus.
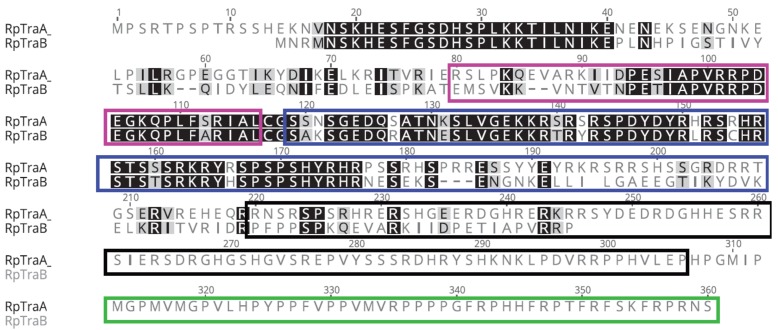
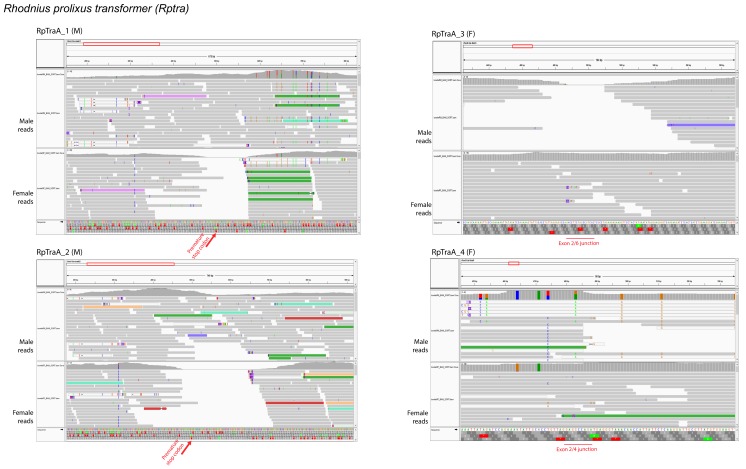
Schematics showing transformer transcripts isolated from R. prolixus, B. germanica, and P. humanus. Coding sequences are in gray, UTRs are in white. All genes are shown as transcribed from left to right. (A) Four transcripts isolated from RpTraA with 5’ and 3’ RACE and confirmed by RNA-seq. Sex-specificity of each transcript is noted in parentheses at the end of the transcript name. Stop codon in the male-specific 3rd exon is indicated with ‘STOP.’ Predicted Transformer protein domains are shown with colored boxes and labels, using RpTraA_4 as example. (B) PCR on male and female cDNA with RpTraA primers (RpTra_checkF + RpTra_checkR in Supplementary file 7) indicated by black arrows in (A) confirms the sex-specificity of each transcript; all amplicons were verified by Sanger sequencing. (C) BgTra spans two scaffolds in the B. germanica genome. Genome coordinates are shown at the beginning and end of each scaffold. Both scaffolds continue beyond BgTra; scaffold 1493 is 552 kb and scaffold 7036 is 55 kb. (D) Six BgTra transcripts were recovered in PacBio Isoseq data and verified by exon-specific RT-PCR. BgTra1, BgTra2 and BgTra4 were recovered from the female Isoseq dataset; BgTra3, BgTra5, and BgTra6 were recovered from the male Isoseq dataset; however, PCR and qPCR on male and female cDNA showed the presence of BgTra3-6 in both sexes (E, F). Nucleotides that code for important protein domains are color-coded as in panel A, using BgTra5 and BgTra6 as examples. BgTra1 and BgTra2 have alternative polyadenylation sites, marked with blue text. In BgTra1 and BgTra2, exon six extends ~500 bp further 3’ than in all other transcripts. The 3’ end of exon six in BgTra3-6 falls six bp upstream of the stop codon that ends the conceptual translation of BgTra1 and BgTra2. Exon 8 codes for 47 amino acids that interrupt the CAM domain, which spans the end of exon six and the start of exon 9. Purple lines show the locations of dsRNA used for RNAi knock-down. Black arrows indicate locations of primers used for RT-PCR and qPCR. (E) qPCR expression of different BgTra isoforms. The truncated transcripts BgTra1 and BgTra2 are strongly female-biased, while the remaining transcripts are sexually monomorphic. (F) RT-PCR expression of different BgTra isoforms. Top panel: both males and females express transcripts with and without the exon interrupting the CAM domain. The shorter band contains BgTra isoforms that skip exon eight and contain an intact CAM domain; the larger product contains exons 6, 8, and 9. Middle panel: both males and females express exon 7, found only in BgTra3 and producing a truncated transcript. Bottom panel: both males and females express the extended exon three with a premature stop codon found in BgTra4. (G) Four PhTra transcripts isolated from P. humanus by 5’ and 3’ RACE. PhTra3 has a premature stop codon and is found in both sexes, as shown by RT-PCR on male and female cDNA in panel (H). Forward primer for PCR shown in (H) spanned the exon 5/7 junction (marked with black arrows in panel (G)); reverse primer for this PCR is in exon 7 (also noted with a black arrow). All PhTra isoforms contain the ‘interrupting exon’ inside of the CAM domain. Double bars on scaffold in panel (G) represent breaks in scaffold scale.