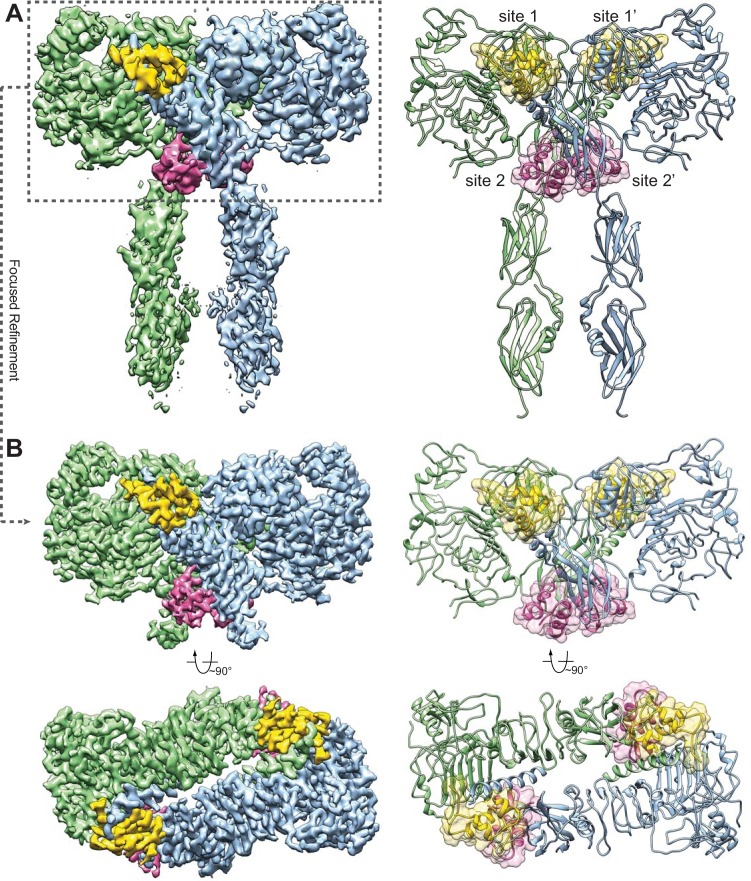
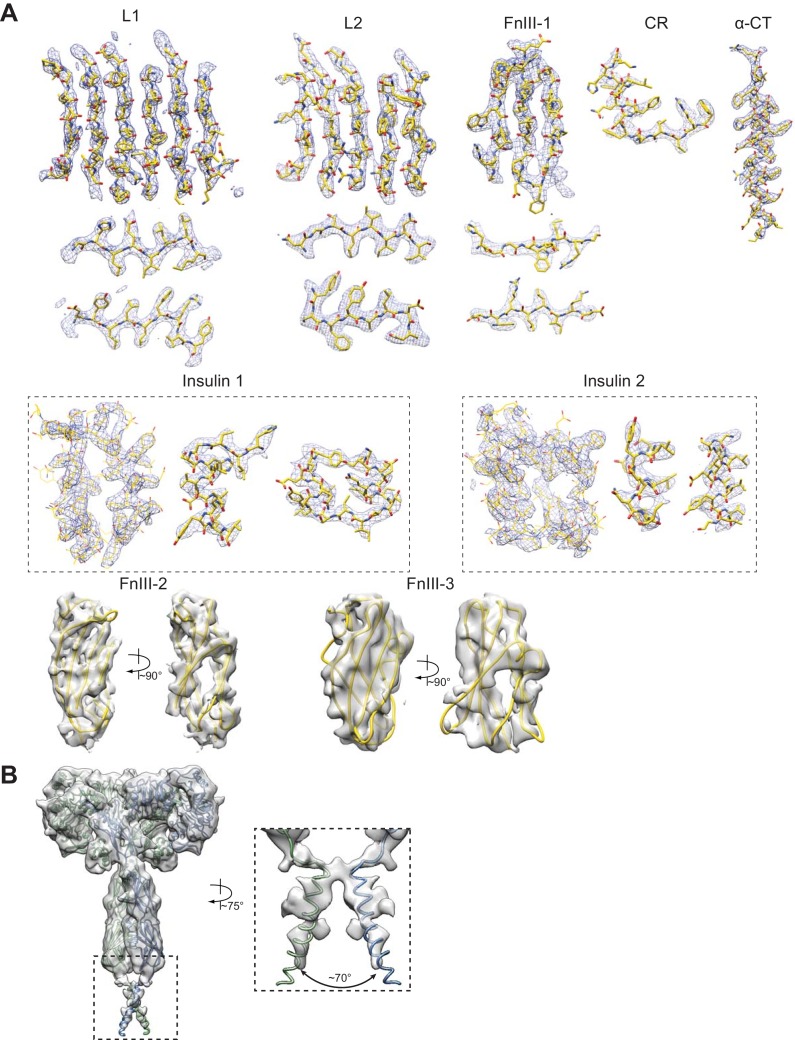
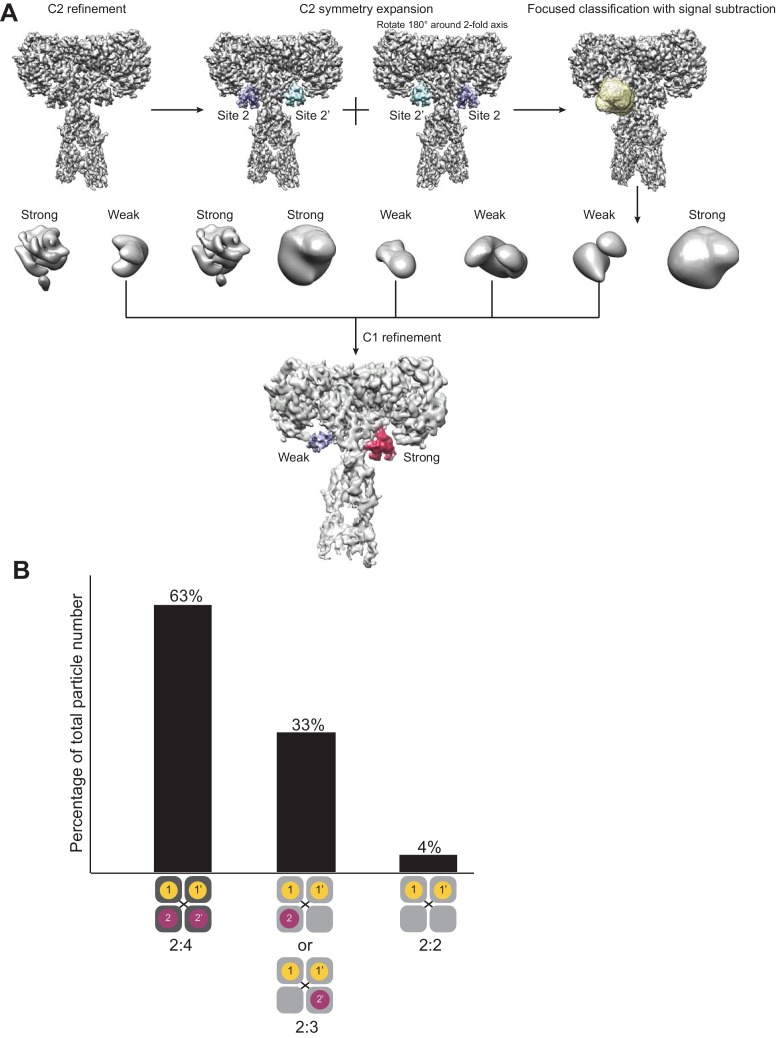
Figure 1. Overall structure of the IR–insulin complex.
(A) 3D reconstruction of the IR dimer with four insulins bound (left) and the corresponding ribbon representation of this complex (right). (B) 3D reconstruction of IR dimer with four insulins bound after forced 3D refinement (left) and the corresponding ribbon representation (right).