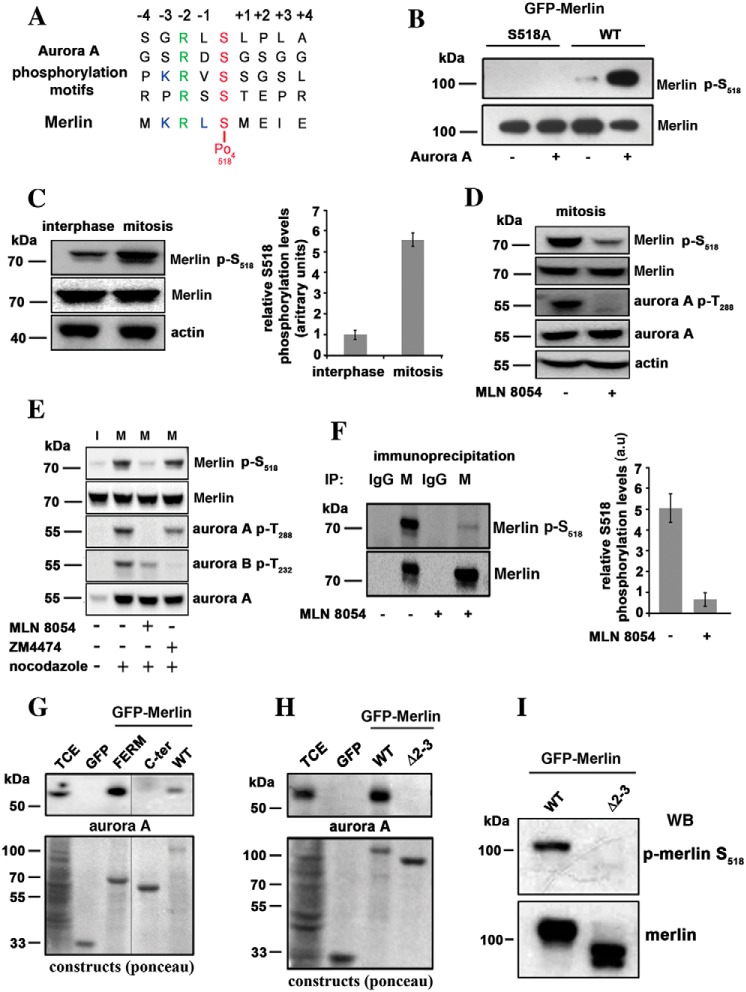
Figure 1.
Merlin is a substrate for Aurora A on serine 518 during mitosis. A, the sequence of amino acids around serine 518 closely matches Aurora A target sequences. B, GFP-Merlin (WT) and GFP-Merlin 518A were immunopurified from HEK293, dephosphorylated with alkaline phosphatase, and incubated with recombinant Aurora A (+) or with the kinase buffer alone (−). The phosphorylation of Ser-518 was detected using an antibody specific to the phosphorylated site. C, the phosphorylation of Ser-518 is higher in extracts from mitotic HeLa cells overexpressing Merlin compared with interphasic cells. The quantification of three independent experiments is shown on the right graph (a.u = arbitrary units). D, mitotic HeLa cells overexpressing Merlin were treated with the Aurora A inhibitor MLN8054. The phosphorylation of Merlin on Ser-518 and the activity of Aurora A (phosphorylation of Thr-288) are strongly inhibited. E, mitotic (+ nocodazole) HeLa cells overexpressing Merlin (M) were treated with Aurora A inhibitor (MLN8054) or Aurora B inhibitor (ZM4474) in the presence of MG132 proteasome inhibitor. Only MLN8054 inhibited phosphorylation on Ser-518. Interphasic cell extract (I) is presented for comparison (− nocodazole). F, endogenous Merlin (M) was immunoprecipitated from extracts of mitotic HEK293 cells treated or not with MLN8054. The samples were probed with antibodies to Merlin or p-Merlin Ser-518. Nonspecific antibody was use as a negative control (IgG). Quantification is presented on the right graph (three experiments). G, co-immunoprecipitation using GFP fusion of Merlin full-length (WT), Merlin FERM domain (FERM, 1–330) or Merlin C-terminal half (330–595, C-terminal) demonstrate that Merlin interacts with Aurora A via its FERM domain. TCE, total cell extract. H, a patient mutant of Merlin FERM domain skipping exons 2 and 3 (Δ2–3, amino acids 38 to 121) fails to bind to Aurora A as observed by co-immunoprecipitation. I, Δ2–3 mutant is not phosphorylated on Ser-518 when expressed in mitotic HeLa cells. Actin was used as loading control throughout. All blots shown above are representative of three independent biological experiments (n = 3). Error bars represent mean ± S.D.