Abstract
Pleiotropic drug resistance arises by the enhanced extrusion of bioactive molecules and is present in a wide range of organisms, ranging from fungi to human cells. A key feature of this adaptation is the sensitive detection of intracellular xenobiotics by transcriptional activators, activating expression of multiple drug exporters. Here, we investigated the selectivity and sensitivity of the budding yeast (Saccharomyces cerevisiae) multidrug response to better understand how differential drug recognition leads to specific activation of drug exporter genes and to drug resistance. Applying live-cell luciferase reporters, we demonstrate that the SNQ2, PDR5, PDR15, and YOR1 transporter genes respond to different mycotoxins, menadione, and hydrogen peroxide in a distinguishable manner and with characteristic amplitudes, dynamics, and sensitivities. These responses correlated with differential sensitivities of the respective transporter mutants to the specific xenobiotics. We further establish a binary vector system, enabling quantitative determination of xenobiotic–transcription factor (TF) interactions in real time. Applying this system we found that the TFs Pdr1, Pdr3, Yrr1, Stb5, and Pdr8 have largely different drug recognition patterns. We noted that Pdr1 is the most promiscuous activator, whereas Yrr1 and Stb5 are selective for ochratoxin A and hydrogen peroxide, respectively. We also show that Pdr1 is rapidly degraded after xenobiotic exposure, which leads to a desensitization of the Pdr1-specific response upon repeated activation. The findings of our work indicate that in the yeast multidrug system, several transcriptional activators with distinguishable selectivities trigger differential activation of the transporter genes.
Keywords: stress response, multidrug transporter, xenobiotic, gene transcription, Saccharomyces cerevisiae, transcription factor, drug resistance, ABC transporter, chemical stress, efflux pump
Introduction
Eukaryotic cells can resist diverse toxic compounds by the action of pleiotropic membrane transporters. These transport proteins, located at the plasma membrane and other intracellular membranes, belong to the ubiquitous ATP-binding cassette (ABC) superfamily (1, 2). The biological functions of ATP transporters from yeast to human cells are numerous. They affect, for example, resistance to cytotoxic compounds with special relevance for chemotherapy in cancer cells, herbicide tolerance in plants or antifungal resistance in pathogenic yeasts (3–5), but as well intracellular homeostatic pathways (6). Also in budding yeast, ATP transporters represent a very numerous protein family, which contains a subgroup of pleiotropic drug resistance (PDR)3 pumps capable of the active extrusion of antifungal toxins from the cytosol (4). This transport activity consumes ATP and is normally repressed in the absence of xenobiotic molecules. An important regulation occurs at the level of transcription at the respective transporter-encoding genes, which is usually highly activated in response to specific xenobiotic compounds (7). Constitutive overexpression of drug exporters is the primary mechanism for cells to acquire pleiotropic drug resistance (4, 8, 9). Gain of function mutations in the transcriptional activator Pdr1, for example, are often found in clinical isolates of Candida species with hyperresistance to antifungal azole drugs (10, 11).
In budding yeast, specialized members of the zinc-cluster transcription factor family orchestrate xenobiotic-induced gene expression. The main activators are the orthologous Pdr1 and Pdr3 proteins (12–14). However, other family members, such as Pdr8, Yrm1, Yrr1, and Stb5, have additional and sometimes repressive functions in gene regulation upon xenobiotic stress (15–21). The mechanisms underlying gene activation upon xenobiotic exposure have been investigated mainly for the Pdr1 and Pdr3 factors. Activation occurs directly through recognition of the bioactive compound by a specialized xenobiotic-binding domain (XBD) in Pdr1 and Pdr3 (22). Both transcription factors seem to bind constitutively, even in the absence of xenobiotics, to pleiotropic drug resistance elements (PDRE) located at the upstream control regions of drug efflux genes (23). Thus, the critical event of xenobiotic-induced gene expression is the binding of the compound to the Pdr transcription factor at the chromatin of the responsive transporter genes. This event eventually triggers a conformational change in the transcription factor, which activates its C-terminal transactivation domain and allows the recruitment of general co-activator complexes. A direct contact with the Mediator subunit Gal11 has been identified with an important role in this activation process (22, 24, 25). However, other co-activators, such as SWI/SNF, SAGA, FACT, and Rpd3, have been involved in Pdr1-mediated transcriptional control (23, 26, 27). It is important to note that xenobiotic-stimulated gene expression is a rapid and transient process (27), which presumably depends on the nature and dose of the toxic compound.
We have previously developed a real-time reporter system based on the expression of very short-lived luciferase in yeast cells, which quantifies in parallel the dose-dependent activation of gene expression over a wide range of stressor concentrations (28). This system reveals complete dose-response profiles for a given gene and stressor in the living cell and thus can distinguish sensitivities of stress-activated transcription factors or gene promoters (29, 30). Here we investigated the yeast multidrug response as a dose-dependent biological system and applied destabilized luciferase reporters to discern the sensitivities of the participating transcription factors and the up-regulated multidrug transporter genes.
Results
Differential regulation of multidrug transporter genes in response to xenobiotics
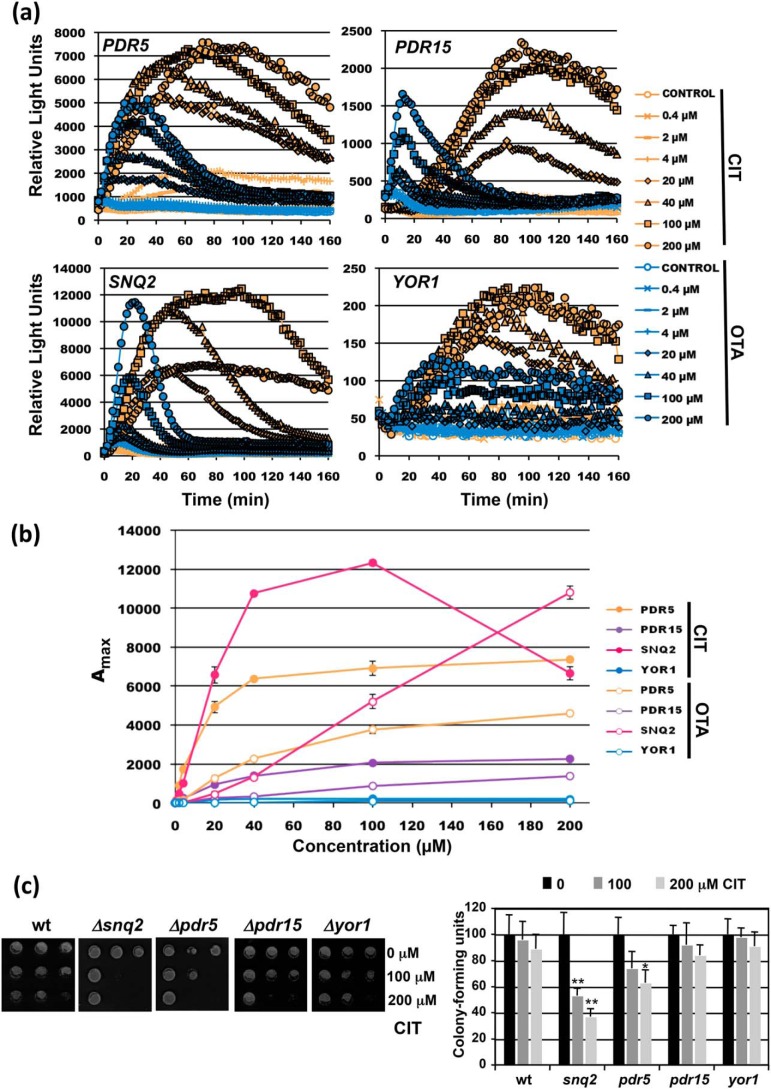
To quantify the dose-dependent induction of transporter-encoding genes in response to xenobiotics, we created live-cell yeast reporter strains by genomic replacement of PDR5, SNQ2, PDR15, and YOR1 with destabilized luciferase (lucCP+) as described previously (30). The engineered strains permit the time-elapsed visualization of gene expression triggered by the four different natural promoter regions. We have previously found that certain mycotoxins activate the yeast multidrug response differentially (31). Therefore, we first compared the expression profiles of the four genes upon the exposure to increasing doses of the mycotoxins citrinin (CIT) and ochratoxin A (OTA). As shown in Fig. 1a, all reporter strains were suitable to obtain complete dose-response profiles on the mycotoxin treatments. Moreover, we observed different induction patterns and dynamics comparing the four representative transporter genes. Rapid and efficient induction by both toxins was observed at PDR5, whereas YOR1 responded much slower and less efficiently. SNQ2 and PDR15 were induced more rapidly by OTA as compared with CIT, and generally we observed more transient gene activation by OTA. We then compared the sensitivities of the dose responses for the four promoters by plotting the maximal reporter activity versus the mycotoxin dose (Fig. 1b). In this way we found that CIT and OTA activated SNQ2 or PDR15 with different sensitivities, whereas PDR5 and YOR1 responded to both toxins with comparable sensitivities. We confirmed by growth assays (Fig. 1c) that the Snq2 transporter was the major determinant of CIT tolerance, which correlates with the most efficient activation of SNQ2 expression upon CIT exposure. Taken together, the real-time quantification of gene induction revealed that individual members of the multidrug transporters are regulated by xenobiotic compounds with distinguishable dynamics and sensitivity.
Figure 1.
Real-time gene induction patterns upon CIT and OTA exposure. Yeast strains with genomic fusions of the indicated promoters with destabilized luciferase were used. a, dose-response profiles of PDR5, PDR15, SNQ2, and YOR1 upon the treatment with the indicated mycotoxin concentrations. The light emission from three independent culture aliquots was continuously measured. S.D. was <15% throughout the experiment but is not included in the graphs. b, comparison of the sensitivity of gene induction of the same four reporter genes. Shown are the mean values of three independent measurements including S.D. c, sensitivity assays of the indicated transporter mutants. Cells were treated in liquid culture aliquots with the indicated CIT concentrations. Cell growth was then assayed on YPD plates (left panel) and cfu determined (right panel). Significant differences according to the Student's t test are marked (*, p < 0.05; **, p < 0.01).
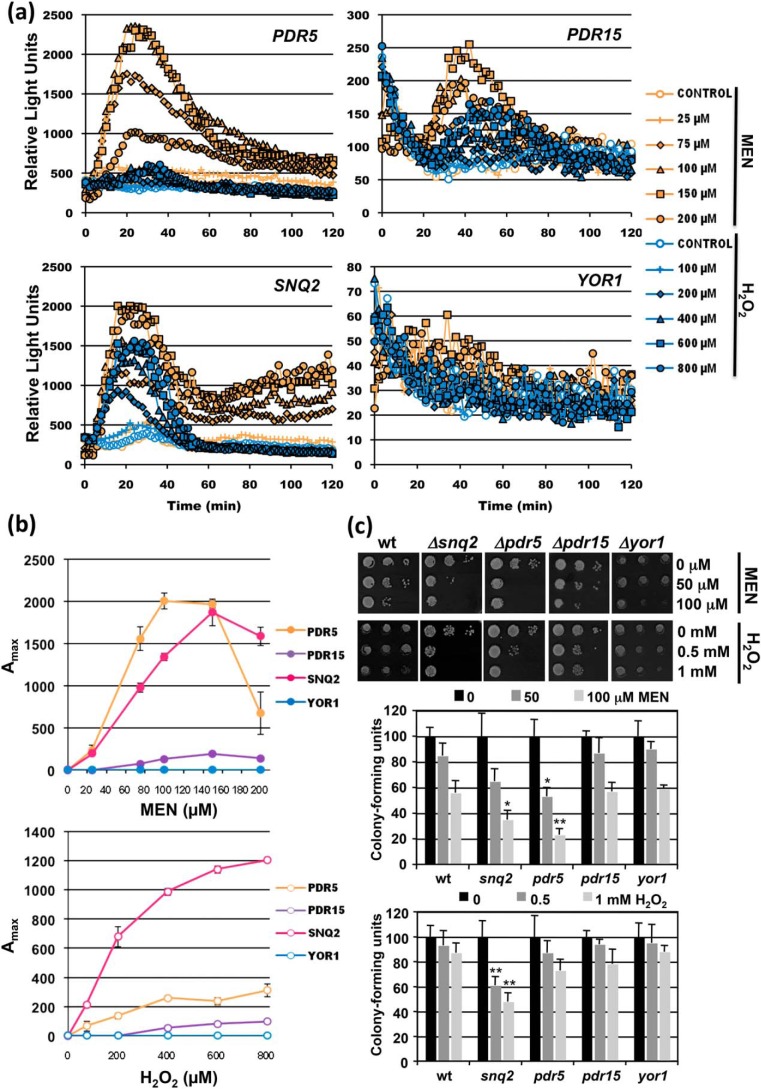
We wanted to expand this finding by the application of different toxic compounds such as the hydrophobic oxidant menadione (MEN) or the hydrophilic, membrane-permeable hydrogen peroxide (H2O2). As summarized in Fig. 2a, only PDR5 and SNQ2 responded robustly to MEN, whereas PDR15 and YOR1 showed very little regulation. Surprisingly, even H2O2, which passes through biological membranes by passive diffusion, highly activates the Snq2 membrane transporter.
Figure 2.
Real-time gene induction patterns upon MEN and H2O2 exposure. The same yeast strains as in Fig. 1 were used. a, dose response profiles of PDR5, PDR15, SNQ2, and YOR1 upon the treatment with the indicated menadione or H2O2 concentrations. The light emission from three independent culture aliquots was continuously measured. S.D. was <15% throughout the experiment but is not included in the graphs. b, comparison of the sensitivity of gene induction of the same four reporter genes. Shown are the mean values of three independent measurements including S.D. c, sensitivity assays of the indicated transporter mutants. Cells were treated in liquid culture aliquots with the indicated MEN and H2O2 concentrations. Cell growth was then assayed on YPD plates (upper panel) and cfu determined (lower panel). Significant differences according to the Student's t test are marked (*, p < 0.05; **, p < 0.01).
The dose-dependent analysis of the promoter activities revealed that the PDR5 and SNQ2 genes had divergent sensitivities toward the two agents (Fig. 2b), which was further confirmed by sensitivity assays (Fig. 2c). We concluded that the four multidrug exporter genes studied here displayed a considerable variability in their dose-dependent activation by the different xenobiotic molecules.
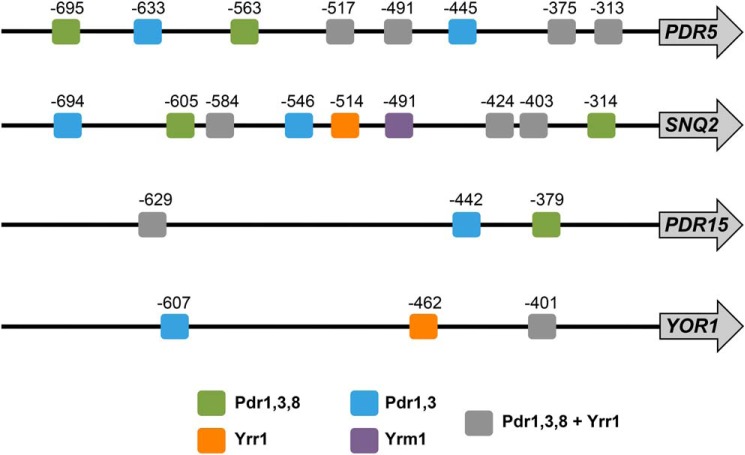
Additional complexity is added by the fact that different multidrug-responsive promoters can be targeted by different Pdr TFs. To determine the promoter variety of different multidrug transporter genes, we mapped potential PDRE sites in the PDR5, SNQ2, PDR15, and YOR1 upstream regulatory sequences. As shown in Fig. 3, the differences in PDRE patterns, both for number and specificity of the elements, is considerable in the four promoters. The presence of many potential PDRE sites in PDR5 and SNQ2 correlates with the high inducibility of these particular genes. Given the complexity of natural genes responding to xenobiotics, we wanted to quantify the specificity of individual transcription factors and their contribution to the multidrug response.
Figure 3.
Distribution of PDRE sequences in multidrug transporter gene promoters. Putative Pdr TF binding sites were identified in the upstream 1000 nucleotides of the indicated genes with the YEASTRACT search machine (46) (www.yeastract.com).4 The consensus sequences are the following: Pdr1/3 = TCCGT/CGG/CA/G; Pdr8 = TCCGHGGA; Yrr1 = WCCGYKKWW; Yrm1 = ACGGAAAT.
A binary vector system for the real-time quantification of compound recognition by the yeast Pdr TFs
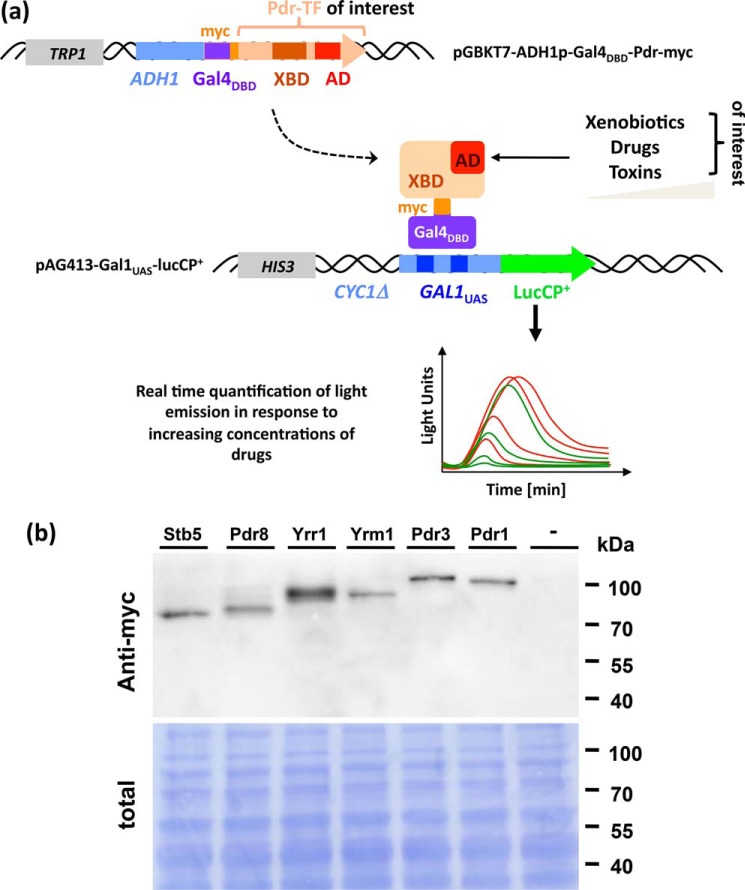
A quantitative analysis of the budding yeast PDR system is complicated by the fact that at least six different TFs are involved in the response to xenobiotic compounds, here referred to as Pdr TFs. Although genetic approaches with null alleles in specific TFs can reveal the physiological relevance of individual factors for specific stresses, the compound selectivity and sensitivity of the individual TFs remain unknown. We wanted to create an experimental setup, which can resolve this question in a quantitative and instantaneous manner. To avoid interference by different DNA binding affinities or in vivo expression levels, we replaced the natural DNA-binding domain of all TFs of the PDR system with Gal4DBD and constitutively expressed the hybrid factors (Fig. 4a). To measure the transcriptional output of the activated TFs in a truly quantitative and time-elapsed manner, we co-transformed the cells with a lucCP+ reporter driven by GAL1UAS sequences (Fig. 4a). We applied this binary system to the yeast Pdr TFs Pdr1, Pdr3, Pdr8, Yrr1, Yrm1, and Stb5, which were stably expressed as Gal4DBD-hybrid proteins (Fig. 4b).
Figure 4.
A binary vector system for the quantitative analysis of Pdr TF activation by xenobiotics in real time. a, plasmid constructions: Hybrid TFs swapping the natural DNA-binding domain of the Pdr factor with Gal4DBD are constitutively expressed from the pGBKT7 vector. The hybrid factor binds to the GAL1UAS sequence of the pAG413 reporter plasmid driving the expression of lucCP+. Dose-dependent activation of the Pdr TF of choice can be monitorized by the time-elapsed detection of light emission. b, expression of six members of the Pdr TF family of yeast. The Gal4DBD-TF fusion proteins were detected by anti-myc Western blotting in whole cell extracts. The total protein content is visualized by DB71 staining and shown below.
Determining the substrate specificities of the yeast Pdr TFs in vivo
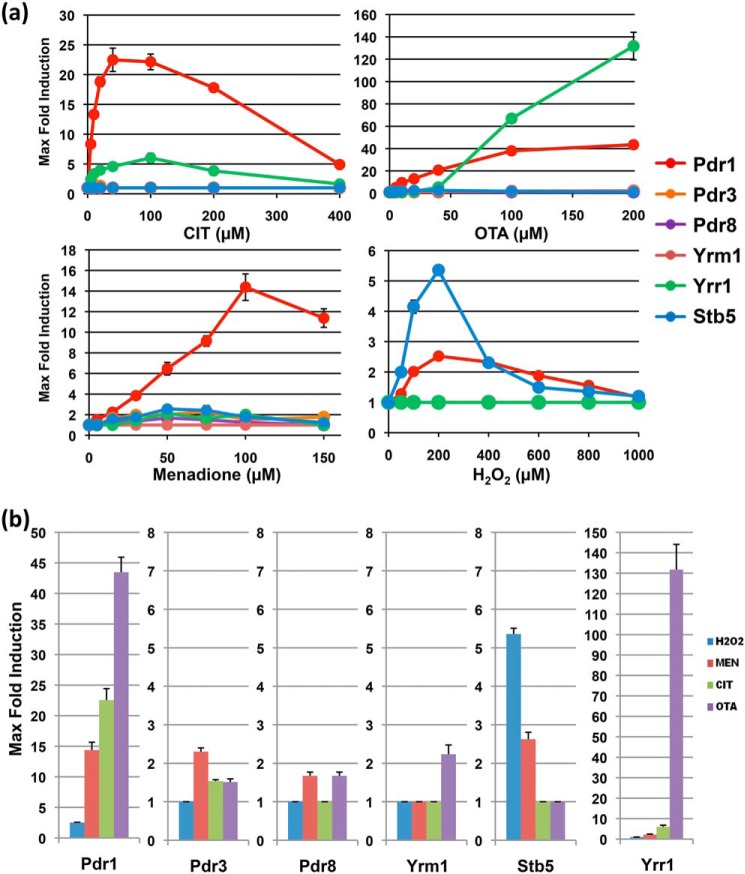
We tested the capacity of each of the Pdr transcription factors to activate gene expression in response to increasing doses of CIT, OTA, MEN, and hydrogen peroxide (Fig. S1). We found that each treatment seemed to activate transcription through more than one specific TF. CIT and OTA were recognized by Pdr1 and Yrr1 with minor contributions of Pdr3 and Yrm1 for OTA. MEN activated mainly Pdr1 but also weakly Stb5, Yrr1, and Pdr3, whereas hydrogen peroxide stimulated mainly Stb5 and to a minor degree Pdr1. Thus the binary quantification system was able to identify specific TF-xenobiotic interaction patterns. In Fig. 5a, we summarize the dose profiles obtained for the six TFs in response to the tested compounds. Interesting differences can be extracted from these profiles. Pdr1, for example, is the TF with the highest affinity for the mycotoxins CIT and OTA. Yrr1 responds to both toxins in a less-sensitive manner, however discriminates much more between the two compounds. Moreover, OTA at high doses activates gene expression through Yrr1 with extraordinary efficiency. Another example of divergent compound activation can be observed for MEN and hydrogen peroxide, both activators of Pdr1 and Stb5. However, Stb5 transactivation increases from MEN to hydrogen peroxide, whereas the opposite is true for Pdr1. In Fig. 5b we represent the different activation patterns observed for the different interventions, which are indicative of differential compound recognition by the six Pdr TFs. Pdr3, Pdr8, and Yrm1 only show 2-fold activation in the best cases, which suggests that the truly interacting compounds have yet to be identified for these TFs or that these specific fusion proteins have impaired function.
Figure 5.
Comparison of compound recognition for the yeast Pdr transcription factors. a, maximal-fold induction plotted versus the compound (citrinin, ochratoxin a, menadione, or H2O2) concentration for the dose-response profiles represented here. b, comparison of the selective inducibility of the Pdr1, Pdr3, Pdr8, Yrm1, Yrr1, and Stb5 TFs by the same compounds. Data are mean values from three independent measurements including S.D.
Stb5 is a high-affinity H2O2-sensing transcriptional activator
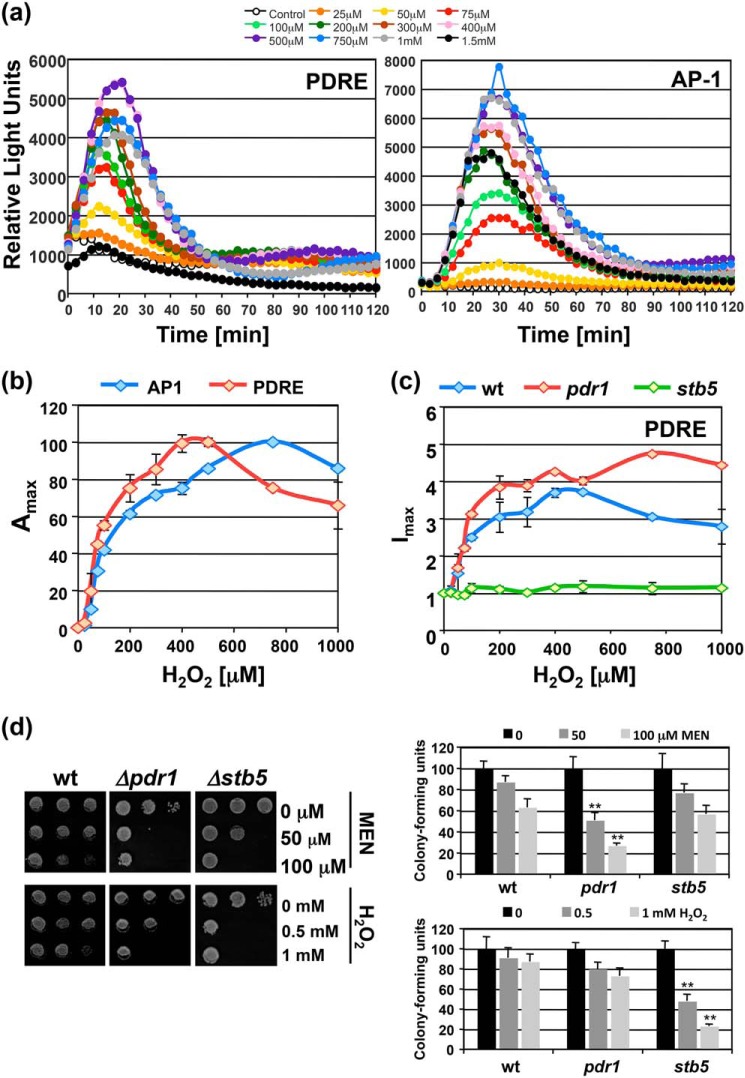
We wanted to characterize in a more quantitative manner the contribution of Stb5 in H2O2 signaling and compare it to the well-known H2O2 sensor Yap1. We used Stb5- and Yap1-specific lucCP+ reporters driven by PDRE- or AP-1–binding sites, respectively, to monitor the H2O2 dose-dependent activation. As shown in Fig. 6a, the application of both reporters yielded the complete hydrogen peroxide–response profile. The analysis of the dose dependence revealed that activation from PDRE sites occurred with higher sensitivity as compared with AP-1 (Fig. 6b). To exclude the interference of other PDRE-binding factors, we tested whether the H2O2 response was mediated by Stb5. As shown in Fig. 6c, we confirmed that an stb5 null allele strain completely lost the peroxide response, whereas a pdr1 mutant was not affected. These data indicated that Stb5 mediates H2O2-responsive transcriptional activation from PDRE sites in a more sensitive fashion than Yap1. Growth assays further confirmed the greater importance of Stb5 in hydrogen peroxide tolerance and Pdr1 in MEN tolerance according to the here-identified specificities of the two transcription factors (Fig. 6d).
Figure 6.
Sensitive H2O2 signal transduction via Stb5. a, yeast WT cells transformed with PDRE- or AP-1–lucCP+ reporters were subjected to the indicated H2O2 concentrations and the gene expression continuously measured by the light emission from three independent culture aliquots for each condition. S.D. was <15% throughout the experiment, but is not included in the graphs. b, comparison of the dose-dependent gene induction by H2O2 via AP-1 or PDRE sites. Maximal luciferase activity plotted versus the H2O2 concentration for the dose-response profiles in (a). c, activation from PDRE sites by H2O2 depends on Stb5. The dose-response profile of PDRE-lucCP+ containing cells of the indicated genetic background was measured and the maximal-fold induction calculated as described in “Experimental procedures.” Data are mean values from three independent measurements including S.D. d, sensitivity assays of the pdr1 and stb5 mutants. Cells were treated in liquid culture aliquots with the indicated MEN and H2O2 concentrations. Cell growth was then assayed on YPD plates (left panel) and cfu determined (right panel). Significant differences according to the Student's t test are marked (**, p < 0.01).
Regulatory function of the PDR transcription factor family upon CIT and OTA exposure
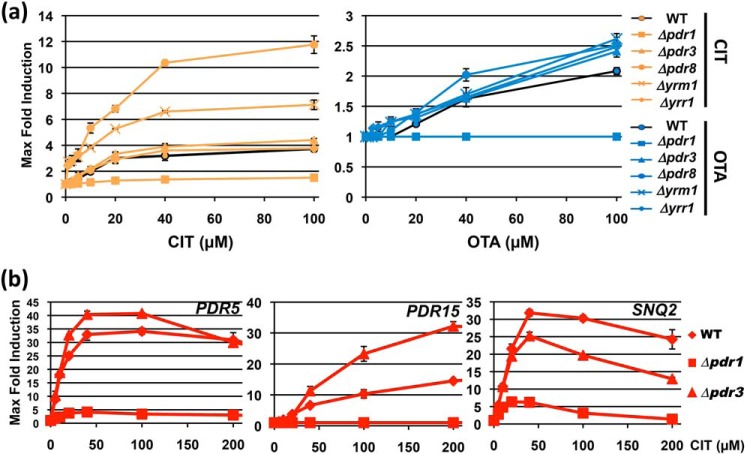
Having characterized some specific interactions between xenobiotics and individual TFs, we wanted to confirm the importance of this selectivity for gene activation upon some compound treatments. The function of all PDR TFs in CIT- and OTA-induced gene expression was assessed. We employed a generic luciferase reporter, which contained the lucCP+ gene controlled by several upstream repeats of the canonical PDRE TF–binding motif. We compared the WT response profile with loss of function mutants in all Pdr TFs (Fig. S2). We confirmed that Pdr1 was the main transcriptional activator operating upon CIT and OTA stress. No other TF seemed to have a positive function in the stimulated expression from PDRE sites (Fig. 7a). Instead we identified Pdr8 and Yrm1 as inhibitors of the mycotoxin-induced gene expression. This was especially detectable for the CIT response, which became faster and more efficient in the respective mutants.
Figure 7.
Functions of the yeast Pdr TFs in the activated gene expression upon CIT and OTA exposure. The indicated yeast strains were assayed with a PDRE-lucCP+ live-cell reporter and the dose-response profiles determined upon CIT and OTA treatment (Fig. S2). a, maximal-fold induction plotted versus the toxin concentration for the dose-response profiles. b, function of Pdr1 and Pdr3 in the activation of multidrug transporter genes by CIT. Yeast strains with the indicated genomic luciferase fusions were used in WT or the pdr1 and pdr3 deletion mutants. The dose-response profiles of PDR5, PDR15, and SNQ2 upon the indicated CIT treatments concentrations were recorded and the maximal-fold induction plotted versus the toxin concentration. Data are mean values from three independent measurements including S.D.
Although considered redundant, we found that Pdr1 and Pdr3 had nonredundant roles in mycotoxin signaling. To exclude artifacts derived from the use of a generic PDRE sequence, we measured the dose-dependent induction of three different natural genes, PDR5, PDR15, and SNQ2, upon CIT exposure in the presence or absence of Pdr1 or Pdr3. As shown in Fig. 7b, we confirmed the divergent functions of both TFs. Pdr1 was completely necessary for the induction of PDR15 and for most of the activation at the PDR5 and SNQ2 genes. Loss of Pdr3 only partially reduced SNQ2 expression at high CIT doses. At PDR5 and especially at PDR15, we observed a more efficient activation of gene expression in the pdr3 mutant. These results suggested that Pdr1 was the main, albeit not the exclusive, activator upon CIT exposure and that Pdr3 was not involved in CIT-induced gene expression but rather displayed an inhibitory function at some target genes. Taken together, the TFs of the PDR family seemed to have divergent positive and negative effects on the toxin-induced gene expression.
Desensibilization of the Pdr1 response after previous substrate recognition
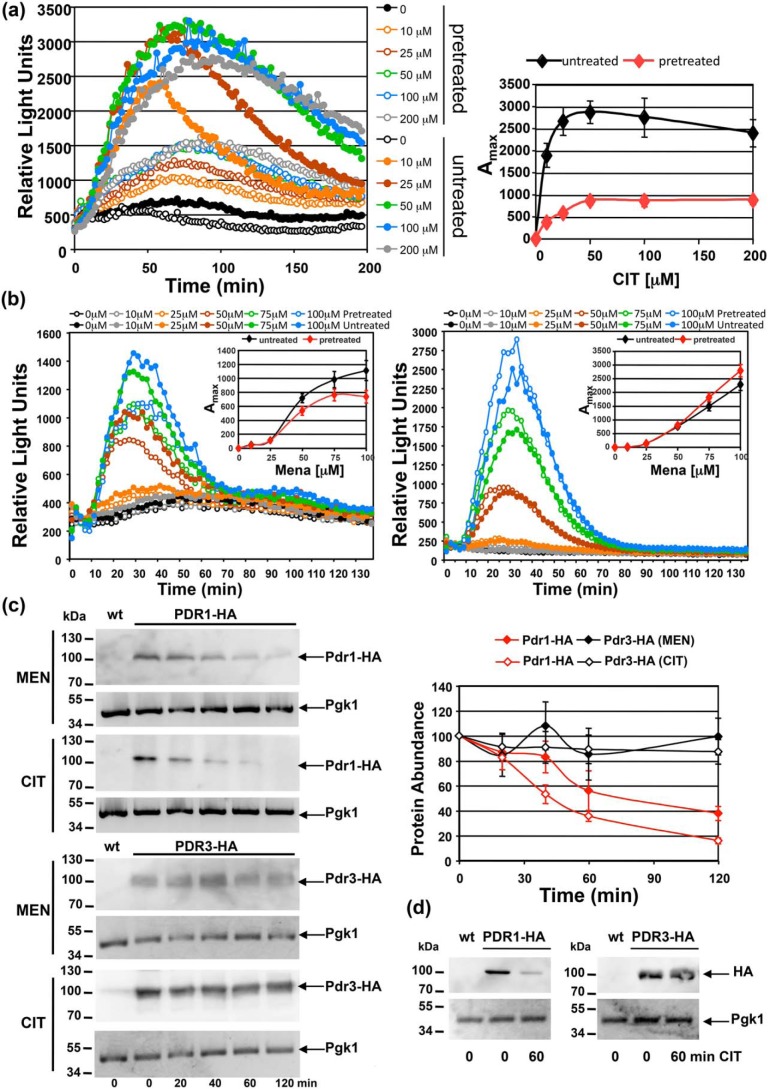
We next wanted to explore how the multidrug response changed after repeated exposure to a particular xenobiotic molecule. We compared the dose response to CIT in cells shortly after a previous CIT exposure with cells that had never been treated with the mycotoxin. As shown in Fig. 8a, we unexpectedly observed that the pretreated cells responded with a significantly lower efficiency to the second CIT stimulus at all concentrations tested. This was not due to a loss of viability during the pretreatment because the CIT concentrations applied did not affect the number of viable cells during the experiments. We next compared this behavior to another TF, Yap1, which responds to some compounds recognized by Pdr1. We used MEN as a stimulus and measured the dose response with the Pdr1-specific PDRE-lucCP+ or with the Yap1-specific AP-1–lucCP+ reporter. We again observed a general decrease of the transcriptional response in the case of Pdr1 (Fig. 8b); however, the effect was less pronounced as compared with CIT. The Yap1 response changed in the opposite way as it slightly gained efficiency after the MEN treatment (Fig. 8b). Pdr1, and not Pdr3, responds to MEN or CIT according to our previous results. Therefore we quantified the Pdr1 protein levels before and during the response to these xenobiotics. We found that the amount of Pdr1 declines rapidly after MEN and CIT exposure, whereas Pdr3 protein levels remained constant (Fig. 8c). We finally tested the stability of Pdr1 and Pdr3 before and after CIT treatment in the presence of cycloheximide (Fig. 8d) and found that levels of Pdr1, but not Pdr3, declined by the CIT exposure. This indicated that Pdr1 was specifically degraded shortly after compound recognition and transcriptional activation. The reduced Pdr1 level might lead to the observed desensibilization of the multidrug response after a previous xenobiotic encounter.
Figure 8.
Previous stimulation reduces Pdr1 protein levels and subsequent gene activation. a, WT yeast cells with a PDRE-lucCP+ reporter were pretreated or not with 50 μm CIT and then subjected to the indicated CIT concentrations. Dose-response curves are shown at the left. The light emission from three independent culture aliquots was continuously measured. S.D. was <15% throughout the experiment but is not included in the graphs. The maximal activity for each CIT concentration is indicated at the right for untreated and pretreated cells. Data are mean values of three independent measurements including S.D. b, same pretreatment experiment as in (a) but comparing PDRE-lucCP+ (left) with AP-1-lucCP+ (right) upon MEN exposure. Cells were pretreated or not with 50 μm MEN. The maximal activity for each MEN concentration is given in the insets. Data are mean values from three independent samples including S.D. c, Pdr1 protein levels specifically decrease upon CIT or MEN exposure. Cells harboring genomic PDR1-HA or PDR3-HA fusions were exposed to 50 μm CIT or MEN for the indicated times. Pdr1 and Pdr3 protein levels were visualized by anti-HA Western blotting in total protein extracts (left). The relative protein abundance is shown at the right. Data are mean values from three independent blots including S.D. d, protein stability was assayed in the presence of 250 μg/ml cycloheximide in Pdr1- and Pdr3-HA expressing cells with or without 50 μm CIT.
Discussion
Our study presents a quantitative approach to understanding the compound-specific activation of gene expression in the yeast multidrug system. One of the first events in the activation of the multidrug extrusion system is the specific recognition of the potentially harmful compound within the cell. In yeast, specialized transcriptional activators act in this forefront of xenobiotic defense by directly binding to the chemical and subsequently activating gene transcription. Here we provide a sensitive in vivo tool to quantify compound-specific transactivation for single TFs of interest based on real-time reporters. Importantly, the xenobiotic-induced transcription is the critical step in the acquisition of drug resistance because resistant isolates of pathogenic yeasts often show constitutive and compound-independent transactivation (10, 32, 33). By coupling the TF activity to a highly quantitative and dose-responsive readout such as the luciferase activity in vivo, the sensitivity of individual TFs toward different chemical compounds can be determined. The measured transactivation very likely reflects the affinity of the TF to the specific compound. Direct binding assays have been performed with the yeast Pdr1 and Pdr3 proteins in vitro, indicating that both TFs are able to bind structurally diverse compounds such as antifungal azoles, cycloheximide or rifampicin (22). Moreover, drug recognition occurs at a discrete XBD in the central part of the proteins. Although these data initially indicated a redundant function of Pdr1 and Pdr3 in the multidrug response, here we demonstrate that the in vivo specificities of both factors are divergent. Pdr1 activates transcription in response to and very likely directly recognizing menadione, citrinin, and ochratoxin A, although none of these compounds is able to efficiently activate Pdr3. Pdr1 and Pdr3 share the highest degree of sequence conservation of the six Pdr-like TFs in yeast, but even in this case their XBDs are only 37% similar. This suggests that different Pdr TFs have acquired specific recognition patterns during evolution. Nonoverlapping functions for Pdr1 and Pdr3 have been suggested before for toxic compounds such as aliphatic solvents which require Pdr3 but not Pdr1 for resistance (34). Our approach reported here will help to distinguish the relevant in vivo target compounds for each TF in the future. It is important to note that sequence similarity does not seem to translate into a similar compound recognition pattern, because here we observe efficient transactivation of Pdr1 and Yrr1 by ochratoxin A, although both proteins share much less sequence similarity than Pdr1 and Pdr3. Previous work on the yeast multidrug system made use of gain of function mutations, in particular TFs leading to hyperresistance to various inhibitor treatments. However, these constitutively active Pdr transcription factors, particularly Pdr1, Pdr3, and Yrr1, show drug-independent gene activation of plasma membrane transporters and are therefore useful to understand how pleiotropic resistance is produced at the level of drug extrusion but not at the level of drug recognition (20, 21, 35–37). Our method described here is able to quantify the compound recognition by individual TFs and in principle should open up research into heterologous fungal and higher eukaryotic drug recognizing TFs. An interesting approach could be the use of site-directed mutagenized or naturally evolved XBDs to quantitatively assess possible changes in drug sensitivity and selectivity.
Another question is how the compound recognition leads to efficient transcriptional initiation for each Pdr TF. It has been shown that Pdr1 activation triggers Mediator recruitment via a specific interaction with its Gal11 subunit (22). However, other xenobiotic-induced TFs might induce transcription by other mechanisms, for example Pdr3 interacts with yet another Mediator subunit Med12 (24). Our time-elapsed assays demonstrate additional compound-specific changes in transactivation, because whereas MEN or OTA induce highly transient gene induction, CIT triggers very much prolonged activation of gene expression. Because this is recapitulated by Pdr1-Gal4– or Yrr1-Gal4–specific induction, we suggest that CIT might convert the responsive TFs to much more stable activators than other compounds.
Xenobiotic-induced gene expression is a transient process. Thus, there must be mechanisms which reset the response after the initial activation. Here we find evidence that proteolysis of the activated Pdr1 protein might be responsible for this reset. The interaction of the xenobiotic molecules with the Pdr TFs might be irreversible, so that the active complex might have to be degraded after successful gene activation. Interestingly, the Pdr-related transcription factor War1 shows a very similar behavior after activation by organic acids, which are directly recognized by the TF (38, 39).
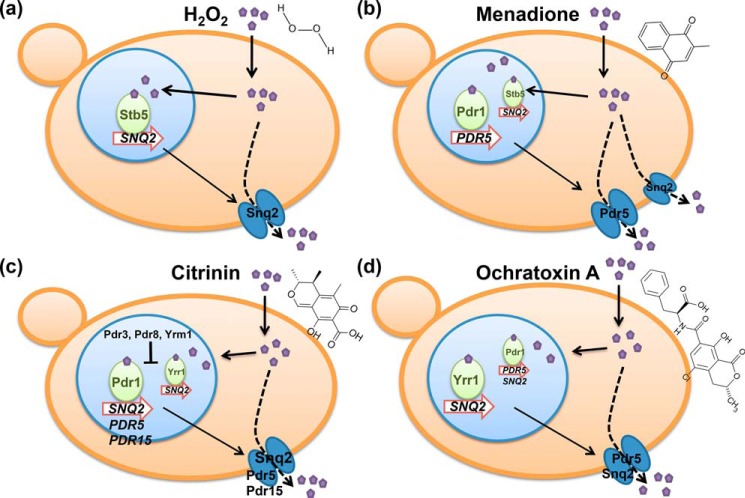
Here we show that different xenobiotic compounds induce the expression of specific multidrug exporters with distinguishable sensitivities. This probably reflects selective adaptation to exclusively express the combination of efflux pumps, which is efficient for the extrusion of the specific compound (Fig. 9). Snq2 and Pdr5 seem to be equally important for CIT detoxification according to their similar dose-dependent induction profile, whereas OTA induction is much more sensitive for Pdr5 as compared with Snq2. Thus, similar compound discrimination found for the Pdr TFs can be found at their target genes, which in this case suggests that CIT/OTA induction depends on Pdr1 at the PDR5 gene and on Yrr1 at the SNQ2 gene. A similar divergence can be observed for the response to the oxidants MEN and hydrogen peroxide at the two transporter genes, most likely produced by the involvement of either Pdr1 or Stb5 in the induction.
Figure 9.
Overview of the differential activation of the yeast multidrug system. a, hydrogen peroxide most sensitively activates Stb5 and the Snq2 transporter. b, menadione targets mainly Pdr1, but also Stb5, with Pdr5 as the most sensitively up-regulated transporter. c, citrinin activates transcription mainly through Pdr1, but also through Yrr1 with less efficiency. Snq2 and Pdr5 are induced with comparable sensitivities. d, ochratoxin A efficiently activates Yrr1 and the Snq2 transporter, however with less sensitivity than Pdr1 and the Pdr5 transporter.
Pdr8 and Yrm1 do not seem to respond significantly to any of the xenobiotics tested here and moreover, cause inhibition for other Pdr activators to induce gene expression from generic PDRE sites. This likely reflects competition of these uninduced factors with the activated factors at the same DNA-binding site rather than generic repression and confirms previous data reporting mutual competition for example between Yrm1 and Yrr1 (15).
We report here a function for the Stb5 protein as a highly sensitive hydrogen peroxide–sensing transcription factor, which confirms previous data confirming a role of Stb5 in the transcriptional control of the yeast antioxidant response (40). Although the exact activation mechanism needs to be identified, it is likely that Stb5 receives the peroxide signal while bound to its target promoters. Interestingly it has been suggested before that Stb5 activation upon oxidative stress occurs through a different mechanism as compared with the thioredoxin-dependent Yap1 (40). Additionally, the Stb5-regulated target genes are different from the Yap1 regulon, as Stb5 seems to target primarily metabolic steps of the pentose phosphate pathway and other NADPH-producing enzymes (40, 41). This suggests that Stb5 might be involved in a metabolic shift toward NADPH production (42), which is activated more sensitively than other antioxidant mechanisms dependent on Yap1. Furthermore the specific induction of the Snq2 plasma membrane transporter upon hydrogen peroxide stress needs to be further investigated to find out what transport activity of Snq2 is responsible for the alleviation of H2O2 stress inside the cell. Our quantitative assay of the yeast multidrug response has revealed how different xenobiotics induce specific responses at multidrug exporters and we develop a real-time methodology for the determination of xenobiotic-specific signaling, which should be stimulating for future research covering more bioactive compounds and drug exporters.
Experimental procedures
Yeast strains and growth conditions
Saccharomyces cerevisiae strains used in this study are shown in Table S1. Yeast strains were grown at 28 °C in synthetic growth medium containing 0.67% yeast nitrogen base, 50 mm succinic acid, pH 3, and 2% dextrose synthetic medium (SD). According to the auxotrophies of each strain, adenine (0.025 g/liter), histidine (0.1 g/liter), leucine (0.1 g/liter), methionine (0.1 g/liter), tryptophan (0.05 g/liter), or uracil (0.025 g/liter) was added. For the luciferase assays, the cells were grown overnight to exponential growth phase and then preincubated with luciferin, as indicated below. Genomic fusions of PDR1 and PDR3 with 3× HA were created according to (43).
Plasmid constructions
Plasmids carrying the constructions of the luciferase reporter fusions and oligonucleotides used in this study are shown in Tables S2 and S3, respectively. For the construction of the integrative luciferase reporter fusions PDR5-, PDR15-, SNQ2-, and YOR1-lucCP+, we employed pUG6-lucCP+-CYC1T-KAN as DNA template to amplify the lucCP+-KanMX cassette with gene-specific primers by PCR. The amplified fragment was integrated in the genome by replacing the ORF of the target gene. For the construction of the PDRE-dependent luciferase reporter we used synthetic double-stranded oligonucleotides with BspEI compatible ends (TF-binding sites underlined): CCGGCGATATCTCCGCGGATAGAATACATCCGCGGATCGCGATCATCCGCGGAT. These oligonucleotides were inserted into the BspEI site of plasmid pAG413-CYC1Δ-lucCP+ (28). The binary system developed in this study is composed of two plasmids. For construction of the destabilized luciferase reporter controlled by the upstream activation sequences (UAS) of the GAL1 promoter, we amplified GAL1 (−551/−336) with terminal MunI and BspEI restriction sites. This sequence was inserted into pAG413-CYC1Δ-lucCP+. pGBKT7-ADH1p-Gal4DBD-myc was employed to obtain the expression vector pGBKT7-Gal4DBD-Pdr by inserting each Pdr transcription factor sequence without its DNA-binding domain next to the Gal4DBD of pGBKT7. The Pdr TF sequences were obtained by PCR using primers containing restriction sites for NcoI/EcoRI and BamHI in their 5′ ends. All constructions were verified by sequencing. The resulting Gal4DBD-hybrid proteins were Pdr1 (aa 81–1068), Pdr3 (aa 53–976), Pdr8 (aa 74–701), Yrm1 (aa 73–786), Yrr1 (aa 93–810), and Stb5 (aa 59–743).
Sensitivity assays
Fresh overnight yeast cultures in SD medium were adjusted to the same OD and diluted 1:10, 1:100, and 1:1000 in SD medium in multiwell plates. The indicated drug concentrations were added to the cells from appropriate stock solutions. After 6 h the cells were replicated onto YPD plates and growth was monitored after 2–3 days. Alternatively, quantitative colony assays were performed with fresh overnight cultures in SD medium, which were diluted to A600 = 0.1 and treated with the indicated drug concentrations for 3 h. The number of cfu was then determined by plating the cultures onto YPD agar plates at appropriate dilutions.
Real-time luciferase expression assays
Yeast strains containing the indicated luciferase fusion genes were grown to exponential phase in SD supplemented with the appropriate amino acids and adjusted to pH 3.0 with 50 mm succinic acid. Cultures were adjusted to the same cell density and incubated on a roller for 60 min at 28 °C with 0.5 mm luciferin (free acid; Synchem, Felsberg, Germany) from a 10 mm stock prepared in DMSO. The cells were then transferred in 120-μl aliquots to white 96-well plates (Costar), which contained the indicated stressor concentrations. The light emission was immediately measured in a GloMax microplate Luminometer (Promega) in three biological replicates. The light emission was continuously recorded over the indicated time and raw data processed with Microsoft Excel software.
Pretreatment assays
Pretreatment or “memory” experiments were essentially performed as described previously (44). Briefly, cells containing the indicated lucCP+ live-cell reporter were grown overnight in SD-His medium adjusted to pH 3.0. The cultures were divided and one half was treated for 1 h with 50 μm CIT or MEN whereas the other half was mock treated with the same amount of solvent. Cells were then briefly collected by centrifugation, washed once with growth medium, and finally resuspended to identical density in fresh SD-His medium for 90 min with luciferin treatment, as described above, in the last 60 min. The indicated MEN or CIT concentrations were then applied and the continuous dose response recorded comparing pretreated and untreated cells. We confirmed that none of the pretreatment regimes affected cell viability.
Western blotting
The expression levels and integrity of the Gal4-Pdr fusion proteins were determined by anti-myc immunodetection. Yeast cells carrying or not pGBKT7-Gal4DBD-Pdr for each transcription factor were grown in synthetic medium to an A600 = 1. Total protein extracts were obtained by glass bead lysis in buffer A (50 mm Tris/HCl, pH = 7.5, 15 mm EDTA, 2 mm DTT, 0.1% Triton X-100, 150 mm NaCl, 1 mm PMSF) supplemented with protease inhibitors (cOmpleteTM, Mini, EDTA-free Protease Inhibitor Mixture (Roche)) using the Precellys Evolution homogenizer (Bertin Technologies). The extracts were resuspended in 2× Laemmli buffer (120 mm Tris/HCl, pH 6.8, 3% SDS, 40 mm DTT, 4 mm EDTA, 12% saccharose, 0.1 mg/ml bromphenol blue) and heated for 5 min at 95 °C. Proteins were separated by 10% SDS-PAGE and analyzed by immunoblotting on PVDF membranes using an α-myc mouse mAb (Roche; 1:5000) and an α-HRP–mouse secondary antibody (1:10,000). The bands were visualized with AmershamTM ECLTM Prime Western Blotting Detection System (GE Healthcare) and quantified with a Fujifilm LAS-3000 system. DB71 staining of the membranes was used as a loading control (45). Pdr1-HA and Pdr3-HA proteins were detected in whole-cell extracts of yeast strains expressing the fusion proteins from their genomic loci. Cell extracts were prepared as described above and the proteins visualized with an α-HA mouse mAb (Roche; 1:5000). α-Pgk1 mouse mAb (Abcam; 1:5000) was used as a loading control.
Statistical analyses
All live-cell gene expression studies were performed on three independent culture aliquots for each stress dose. The results were processed in Microsoft Excel. The light units were corrected for the absolute cell number in each assay to represent the relative light units for each stress treatment. -Fold induction (FI) results were obtained dividing maximal luciferase activities by the corresponding initial value. Data were represented as the mean value with the corresponding standard deviation. The typical error rate between three different biological replicates was <10%. Error bars are omitted in the representation of dose-response curves to make the graphs clearly visible.
Author contributions
E. V.-P., A. P.-A., and M. P. conceptualization; E. V.-P., C. L.-P., B. A., and M. P. data curation; E. V.-P. and M. P. formal analysis; E. V.-P., C. L.-P., B. A., and M. P. investigation; E. V.-P., A. P.-A., and M. P. writing-review and editing; A. P.-A. and M. P. supervision; A. P.-A. and M. P. writing-original draft; M. P. resources; M. P. funding acquisition; M. P. methodology; M. P. project administration.
Supplementary Material
This work was supported by Ministerio de Economía y Competitividad Grant BFU2016-75792-R. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2 and Tables S1–S3.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- PDR
- pleiotropic drug resistance
- XBD
- xenobiotic-binding domain
- PDRE
- PDR elements
- lucCP+
- destabilized luciferase
- CIT
- citrinin
- OTA
- ochratoxin A
- UAS
- upstream activation sequences
- aa
- amino acid
- SD
- synthetic defined (medium)
- cfu
- colony-forming unit
- YPD
- yeast extract peptone dextrose.
References
- 1. Colabufo N. A., Berardi F., Contino M., Niso M., and Perrone R. (2009) ABC pumps and their role in active drug transport. Curr. Top. Med. Chem. 9, 119–129 10.2174/156802609787521553 [DOI] [PubMed] [Google Scholar]
- 2. Seeger M. A., and van Veen H. W. (2009) Molecular basis of multidrug transport by ABC transporters. Biochim. Biophys. Acta 1794, 725–737 10.1016/j.bbapap.2008.12.004 [DOI] [PubMed] [Google Scholar]
- 3. Stavrovskaya A. A., and Stromskaya T. P. (2008) Transport proteins of the ABC family and multidrug resistance of tumor cells. Biochemistry (Mosc) 73, 592–604 10.1134/S0006297908050118 [DOI] [PubMed] [Google Scholar]
- 4. Prasad R., and Goffeau A. (2012) Yeast ATP-binding cassette transporters conferring multidrug resistance. Annu. Rev. Microbiol. 66, 39–63 10.1146/annurev-micro-092611-150111 [DOI] [PubMed] [Google Scholar]
- 5. Ito H., and Gray W. M. (2006) A gain-of-function mutation in the Arabidopsis pleiotropic drug resistance transporter PDR9 confers resistance to auxinic herbicides. Plant Physiol 142, 63–74 10.1104/pp.106.084533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jungwirth H., and Kuchler K. (2006) Yeast ABC transporters—a tale of sex, stress, drugs and aging. FEBS Lett. 580, 1131–1138 10.1016/j.febslet.2005.12.050 [DOI] [PubMed] [Google Scholar]
- 7. Paul S., and Moye-Rowley W. S. (2014) Multidrug resistance in fungi: Regulation of transporter-encoding gene expression. Front. Physiol. 5, 143 10.3389/fphys.2014.00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Balzi E., and Moye-Rowley W. S. (2018) Unveiling the transcriptional control of pleiotropic drug resistance in Saccharomyces cerevisiae: Contributions of André Goffeau and his group. Yeast 36, 195–200 10.1002/yea.3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cannon R. D., Lamping E., Holmes A. R., Niimi K., Baret P. V., Keniya M. V., Tanabe K., Niimi M., Goffeau A., and Monk B. C. (2009) Efflux-mediated antifungal drug resistance. Clin. Microbiol. Rev. 22, 291–321 10.1128/CMR.00051-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moye-Rowley W. S. (2019) Multiple interfaces control activity of the Candida glabrata Pdr1 transcription factor mediating azole drug resistance. Curr. Genet. 65, 103–108 10.1007/s00294-018-0870-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Noël T. (2012) The cellular and molecular defense mechanisms of the Candida yeasts against azole antifungal drugs. J. Mycol. Méd. 22, 173–178 10.1016/j.mycmed.2012.04.004 [DOI] [PubMed] [Google Scholar]
- 12. Näär A. M., and Thakur J. K. (2009) Nuclear receptor-like transcription factors in fungi. Genes Dev. 23, 419–432 10.1101/gad.1743009 [DOI] [PubMed] [Google Scholar]
- 13. Fardeau V., Lelandais G., Oldfield A., Salin H., Lemoine S., Garcia M., Tanty V., Le Crom S., Jacq C., and Devaux F. (2007) The central role of PDR1 in the foundation of yeast drug resistance. J. Biol. Chem. 282, 5063–5074 10.1074/jbc.M610197200 [DOI] [PubMed] [Google Scholar]
- 14. Katzmann D. J., Burnett P. E., Golin J., Mahé Y., and Moye-Rowley W. S. (1994) Transcriptional control of the yeast PDR5 gene by the PDR3 gene product. Mol. Cell. Biol. 14, 4653–4661 10.1128/MCB.14.7.4653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lucau-Danila A., Delaveau T., Lelandais G., Devaux F., and Jacq C. (2003) Competitive promoter occupancy by two yeast paralogous transcription factors controlling the multidrug resistance phenomenon. J. Biol. Chem. 278, 52641–52650 10.1074/jbc.M309580200 [DOI] [PubMed] [Google Scholar]
- 16. Onda M., Ota K., Chiba T., Sakaki Y., and Ito T. (2004) Analysis of gene network regulating yeast multidrug resistance by artificial activation of transcription factors: Involvement of Pdr3 in salt tolerance. Gene 332, 51–59 10.1016/j.gene.2004.02.003 [DOI] [PubMed] [Google Scholar]
- 17. Akache B., MacPherson S., Sylvain M. A., and Turcotte B. (2004) Complex interplay among regulators of drug resistance genes in Saccharomyces cerevisiae. J. Biol. Chem. 279, 27855–27860 10.1074/jbc.M403487200 [DOI] [PubMed] [Google Scholar]
- 18. Cui Z., Shiraki T., Hirata D., and Miyakawa T. (1998) Yeast gene YRR1, which is required for resistance to 4-nitroquinoline N-oxide, mediates transcriptional activation of the multidrug resistance transporter gene SNQ2. Mol. Microbiol 29, 1307–1315 10.1046/j.1365-2958.1998.01027.x [DOI] [PubMed] [Google Scholar]
- 19. Hikkel I., Lucau-Danila A., Delaveau T., Marc P., Devaux F., and Jacq C. (2003) A general strategy to uncover transcription factor properties identifies a new regulator of drug resistance in yeast. J. Biol. Chem. 278, 11427–11432 10.1074/jbc.M208549200 [DOI] [PubMed] [Google Scholar]
- 20. Le Crom S., Devaux F., Marc P., Zhang X., Moye-Rowley W. S., and Jacq C. (2002) New insights into the pleiotropic drug resistance network from genome-wide characterization of the YRR1 transcription factor regulation system. Mol. Cell. Biol. 22, 2642–2649 10.1128/MCB.22.8.2642-2649.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang X., Cui Z., Miyakawa T., and Moye-Rowley W. S. (2001) Cross-talk between transcriptional regulators of multidrug resistance in Saccharomyces cerevisiae. J. Biol. Chem. 276, 8812–8819 10.1074/jbc.M010686200 [DOI] [PubMed] [Google Scholar]
- 22. Thakur J. K., Arthanari H., Yang F., Pan S. J., Fan X., Breger J., Frueh D. P., Gulshan K., Li D. K., Mylonakis E., Struhl K., Moye-Rowley W. S., Cormack B. P., Wagner G., and Näär A. M. (2008) A nuclear receptor-like pathway regulating multidrug resistance in fungi. Nature 452, 604–609 10.1038/nature06836 [DOI] [PubMed] [Google Scholar]
- 23. Gao C., Wang L., Milgrom E., and Shen W. C. (2004) On the mechanism of constitutive Pdr1 activator-mediated PDR5 transcription in Saccharomyces cerevisiae: Evidence for enhanced recruitment of coactivators and altered nucleosome structures. J. Biol. Chem. 279, 42677–42686 10.1074/jbc.M406363200 [DOI] [PubMed] [Google Scholar]
- 24. Shahi P., Gulshan K., Näär A. M., and Moye-Rowley W. S. (2010) Differential roles of transcriptional Mediator subunits in regulation of multidrug resistance gene expression in Saccharomyces cerevisiae. Mol. Biol. Cell 21, 2469–2482 10.1091/mbc.e09-10-0899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nishikawa J. L., Boeszoermenyi A., Vale-Silva L. A., Torelli R., Posteraro B., Sohn Y. J., Ji F., Gelev V., Sanglard D., Sanguinetti M., Sadreyev R. I., Mukherjee G., Bhyravabhotla J., Buhrlage S. J., Gray N. S., Wagner G., Näär A. M., and Arthanari H. (2016) Inhibiting fungal multidrug resistance by disrupting an activator-Mediator interaction. Nature 530, 485–489 10.1038/nature16963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Borecka-Melkusova S., Kozovska Z., Hikkel I., Dzugasova V., and Subik J. (2008) RPD3 and ROM2 are required for multidrug resistance in Saccharomyces cerevisiae. FEMS Yeast Res. 8, 414–424 10.1111/j.1567-1364.2007.00352.x [DOI] [PubMed] [Google Scholar]
- 27. Voth W. P., Takahata S., Nishikawa J. L., Metcalfe B. M., Näär A. M., and Stillman D. J. (2014) A role for FACT in repopulation of nucleosomes at inducible genes. PLoS One 9, e84092 10.1371/journal.pone.0084092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rienzo A., Pascual-Ahuir A., and Proft M. (2012) The use of a real-time luciferase assay to quantify gene expression dynamics in the living yeast cell. Yeast 29, 219–231 10.1002/yea.2905 [DOI] [PubMed] [Google Scholar]
- 29. Dolz-Edo L., Rienzo A., Poveda-Huertes D., Pascual-Ahuir A., and Proft M. (2013) Deciphering dynamic dose responses of natural promoters and single cis elements upon osmotic and oxidative stress in yeast. Mol. Cell. Biol. 33, 2228–2240 10.1128/MCB.00240-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rienzo A., Poveda-Huertes D., Aydin S., Buchler N. E., Pascual-Ahuir A., and Proft M. (2015) Different mechanisms confer gradual control and memory at nutrient- and stress-regulated genes in yeast. Mol. Cell. Biol. 35, 3669–3683 10.1128/MCB.00729-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vanacloig-Pedros E., Proft M., and Pascual-Ahuir A. (2016) Different toxicity mechanisms for citrinin and ochratoxin A revealed by transcriptomic analysis in yeast. Toxins (Basel) 8, E273 10.3390/toxins8100273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cavalheiro M., Costa C., Silva-Dias A., Miranda I. M., Wang C., Pais P., Pinto S. N., Mil-Homens D., Sato-Okamoto M., Takahashi-Nakaguchi A., Silva R. M., Mira N. P., Fialho A. M., Chibana H., Rodrigues A. G., Butler G., and Teixeira M. C. (2019) A transcriptomics approach to unveiling the mechanisms of in vitro evolution toward fluconazole resistance of a Candida glabrata clinical isolate. Antimicrob. Agents Chemother. 63, e00995 10.1128/AAC.00995-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Healey K. R., and Perlin D. S. (2018) Fungal resistance to echinocandins and the MDR phenomenon in Candida glabrata. J. Fungi (Basel) 4, E105 10.3390/jof4030105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nishida-Aoki N., Mori H., Kuroda K., and Ueda M. (2015) Activation of the mitochondrial signaling pathway in response to organic solvent stress in yeast. Curr. Genet. 61, 153–164 10.1007/s00294-014-0463-9 [DOI] [PubMed] [Google Scholar]
- 35. Keeven J., Ko D., Shallom J., Uccelini B., and Golin J. (2002) PDR2 Gain-of-function mutations eliminate the need for Pdr1 and require the UBP6 product for resistance to translational inhibitors. Curr. Genet. 41, 11–19 10.1007/s00294-002-0274-2 [DOI] [PubMed] [Google Scholar]
- 36. Kodo N., Matsuda T., Doi S., and Munakata H. (2013) Salicylic acid resistance is conferred by a novel YRR1 mutation in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 434, 42–47 10.1016/j.bbrc.2013.03.069 [DOI] [PubMed] [Google Scholar]
- 37. Rong-Mullins X., Ayers M. C., Summers M., and Gallagher J. E. G. (2018) Transcriptional profiling of Saccharomyces cerevisiae reveals the impact of variation of a single transcription factor on differential gene expression in 4NQO, fermentable, and nonfermentable carbon sources. G3 (Bethesda) 8, 607–619 10.1534/g3.117.300138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim M. S., Cho K. H., Park K. H., Jang J., and Hahn J. S. (2018) Activation of Haa1 and War1 transcription factors by differential binding of weak acid anions in Saccharomyces cerevisiae. Nucleic Acids Res. 47, 1211–1224 10.1093/nar/gky1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kren A., Mamnun Y. M., Bauer B. E., Schüller C., Wolfger H., Hatzixanthis K., Mollapour M., Gregori C., Piper P., and Kuchler K. (2003) War1p, a novel transcription factor controlling weak acid stress response in yeast. Mol. Cell. Biol. 23, 1775–1785 10.1128/MCB.23.5.1775-1785.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Larochelle M., Drouin S., Robert F., and Turcotte B. (2006) Oxidative stress-activated zinc cluster protein Stb5 has dual activator/repressor functions required for pentose phosphate pathway regulation and NADPH production. Mol. Cell. Biol. 26, 6690–6701 10.1128/MCB.02450-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cadière A., Galeote V., and Dequin S. (2010) The Saccharomyces cerevisiae zinc factor protein Stb5p is required as a basal regulator of the pentose phosphate pathway. FEMS Yeast Res. 10, 819–827 10.1111/j.1567-1364.2010.00672.x [DOI] [PubMed] [Google Scholar]
- 42. Ouyang L., Holland P., Lu H., Bergenholm D., and Nielsen J. (2018) Integrated analysis of the yeast NADPH-regulator Stb5 reveals distinct differences in NADPH requirements and regulation in different states of yeast metabolism. FEMS Yeast Res. 18, foy091 10.1093/femsyr/foy091 [DOI] [PubMed] [Google Scholar]
- 43. De Antoni A., and Gallwitz D. (2000) A novel multi-purpose cassette for repeated integrative epitope tagging of genes in Saccharomyces cerevisiae. Gene 246, 179–185 10.1016/S0378-1119(00)00083-4 [DOI] [PubMed] [Google Scholar]
- 44. Pascual-Ahuir A., González-Cantó E., Juyoux P., Pable J., Poveda-Huertes D., Saiz-Balbastre S., Squeo S., Ureña-Marco A., Vanacloig-Pedros E., Zaragoza-Infante L., and Proft M. (2019) Dose dependent gene expression is dynamically modulated by the history, physiology and age of yeast cells. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 457–471 10.1016/j.bbagrm.2019.02.009 [DOI] [PubMed] [Google Scholar]
- 45. Hong H. Y., Yoo G. S., and Choi J. K. (2000) Direct Blue 71 staining of proteins bound to blotting membranes. Electrophoresis 21, 841–845 [DOI] [PubMed] [Google Scholar]
- 46. Teixeira M. C., Monteiro P. T., Palma M., Costa C., Godinho C. P., Pais P., Cavalheiro M., Antunes M., Lemos A., Pedreira T., and Sá-Correia I. (2018) YEASTRACT, an upgraded database for the analysis of transcription regulatory networks in Saccharomyces cerevisiae. Nucl. Acids Res. 46, D348–D353 10.1093/nar/gkx842 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.