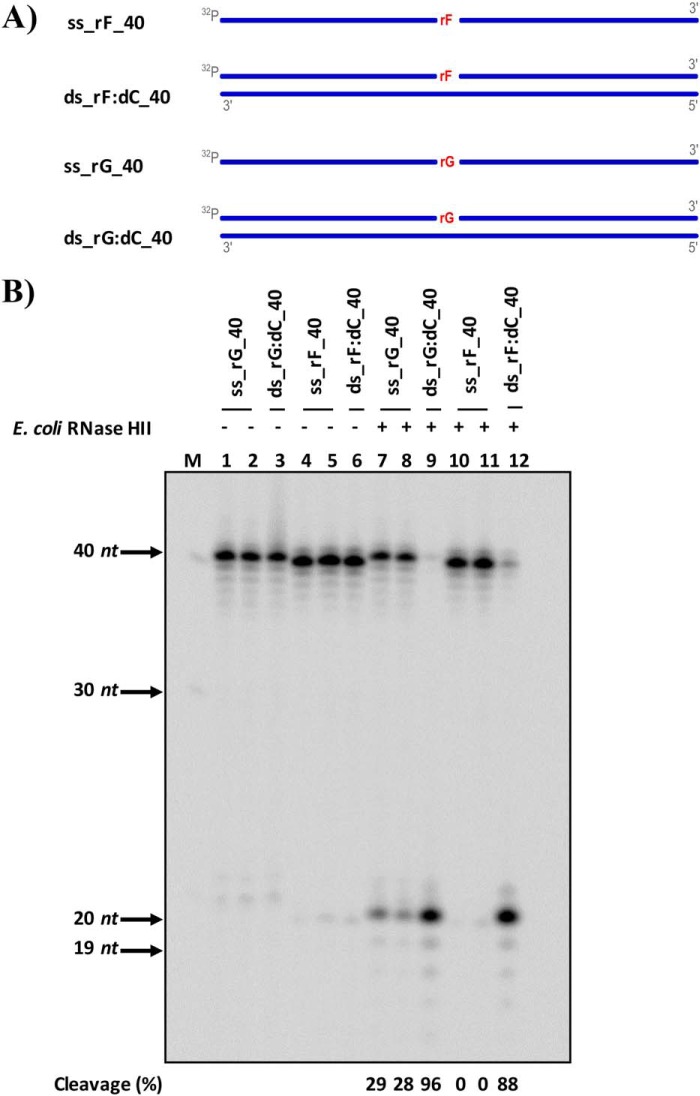
Figure 1.
RNase HII from E. coli cleaves rAP embedded in dsDNA. A, scheme of the ss and ds 40-mer substrates used to test the activity of E. coli RNase HII including a ribo abasic site mimicked by a tetrafuran residue indicated with rF and an unmodified rGMP (rG) embedded in DNA. DNA strands are depicted in blue, whereas rNMPs are in red. The 5′ radiolabels, indicated by 32P, at the 5′ are in gray. B, representative denaturing polyacrylamide gel of cleavage result obtained on all substrates without (lanes 1–6) and with (lanes 7–12) E. coli RNase HII protein. The far left lane, marked with M, corresponds to the ssDNA ladder. On the left side of the image, molecular weights are indicated, with arrows to the corresponding bands. Lanes 1 and 7 and lanes 4 and 10 are ss substrates containing rG and rF, respectively; lanes 2 and 8 and lanes 5 and 11 are ss substrates containing rG and rF, respectively, which are cooled slowly at room temperature to observe any self-annealing; lanes 3 and 9 are ds substrates containing rG; and lanes 6 and 12 are ds substrates containing rF. The cleavage percentages of reactions are displayed below the image.