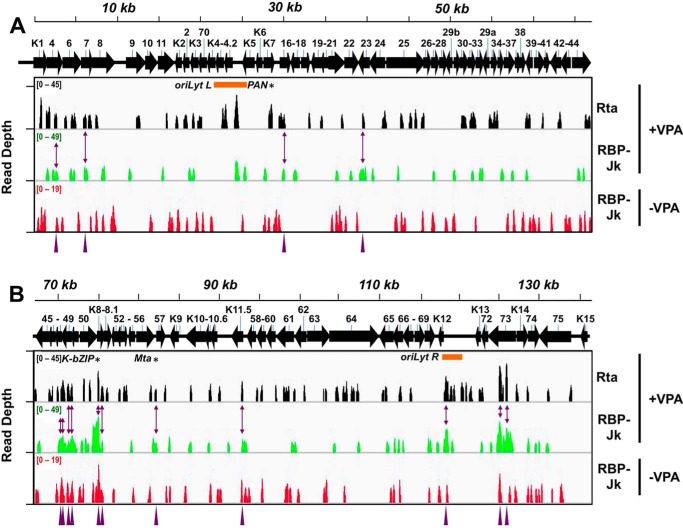
Figure 2.
ChIP/Seq of Rta and RBP-Jκ DNA binding on the KSHV genome during latency and reactivation. Visualization of ChIP/Seq peaks by read depth per bp using Integrative Genomics Viewer (IGV) (85). A, genome positions from bp 1 to 68,000. B, genome positions from bp 68,001 to 137,508. Antibodies used are indicated at right, with chromatin from latent (−VPA) or reactivated (+VPA) virus in BC-3 cells. Solid arrows represent open reading frames (ORFs). Numbering above arrows indicate ORF names. Purple arrowheads and two-headed arrows indicate 15 co-localized Rta and RBP-Jκ peaks in reactivation. Asterisks indicate peak positions mapped to PAN promoter (PAN), ORF50AS/K-bZIP promoter (K-bZIP), and Mta promoter (Mta). Orange bars indicate positions of ori-Lyts.