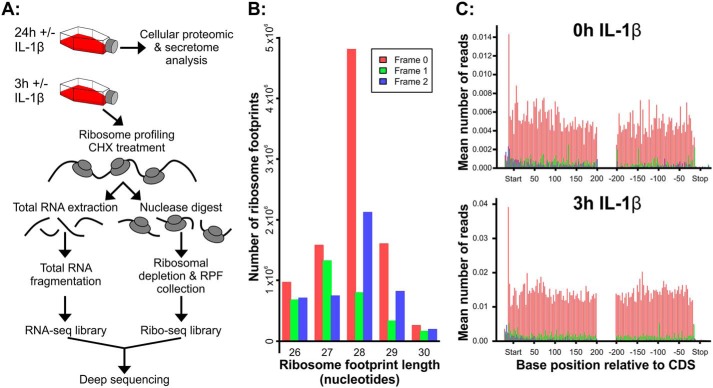
Figure 1.
Ribosome profiling workflow and sequencing data. A, SW1353 workflow. Cells were treated for 3 or 24 h with or without IL-1β. Medium was harvested for secretome proteomic analysis, and cells were harvested for intracellular proteomic analysis after 24-h IL-1β exposure. Ribosome profiling was performed on cells exposed to IL-1β for 0 and 3 h. B, to obtain triplet periodicity of ribosome-protected fragments, sequencing results (FASTQ files) from amplified libraries were first uploaded to RiboGalaxy. The 5′-AGATCGGAAGAGCACACGTCT-3′ adaptor sequence and rRNA sequences were removed using Cutadapt and Bowtie. Sequences were then aligned to the human transcriptome (GRCh38/hg38) using Bowtie prior to riboSeqR analysis. From the riboSeqR analysis, triplet periodicity showed that most sequencing reads were 28 nucleotides in length and in frame 1 (red). C, RiboGalaxy was next used to perform a metagene analysis of the ribosome density and showed similar ribosomal coverage over all transcripts that are 28 nucleotides in length. This representative metagene analysis reflects triplet periodicity results in that the majority of 28-mer transcripts are in frame 1 (red). CHX, cycloheximide.