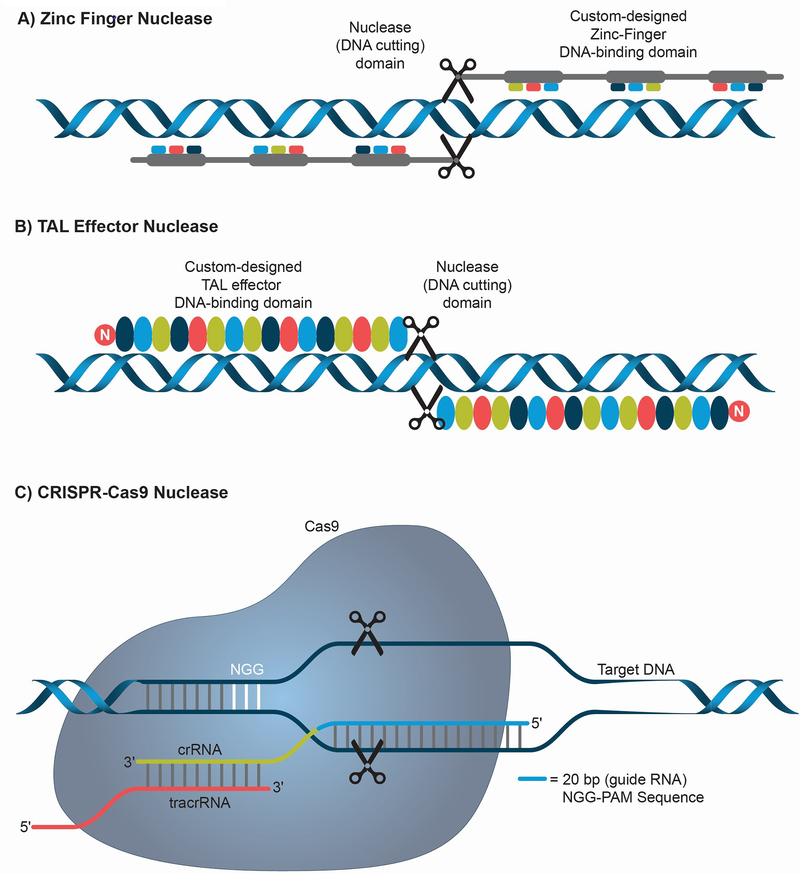
Figure 2: Sequence-specific nucleases used for genome editing.
(A) Zinc Finger Nucleases (ZFN) are artificial restriction enzymes that possess a DNA binding protein domain fused with a nuclease domain to achieve sequence-specific cleavage. ZFNs are designed in pairs to produce double stranded breaks (DSB). (B) TAL Effector Nucleases (TALEN) are restriction enzymes that possess TAL effector DNA binding domains fused to a nuclease domain to achieve sequence-specific cleavage. TALENs are also designed in pairs to enable a highly specific DSB. (C) CRISPR-Cas9 (Clustered Regularly Interspaced Palindromic Repeats-CRISPR associated protein 9) are sequence specific nucleases discovered as part of a bacterial antiviral immune mechanism. A short RNA (guide RNA, usually 20 bp) complementary to the target DNA sequence is utilized to direct the Cas9 protein to a specific genomic region to create DSB. The system requires the presence of a short sequence in the genomic DNA adjacent to the guide RNA target sequence called the protospacer adjacent motif (PAM) which in the case of Streptococcus pyogenes Cas9 is NGG (N can be any nucleotide). crRNA and tracrRNA associate with the Cas9 to help in recognizing and cleaving target DNA.