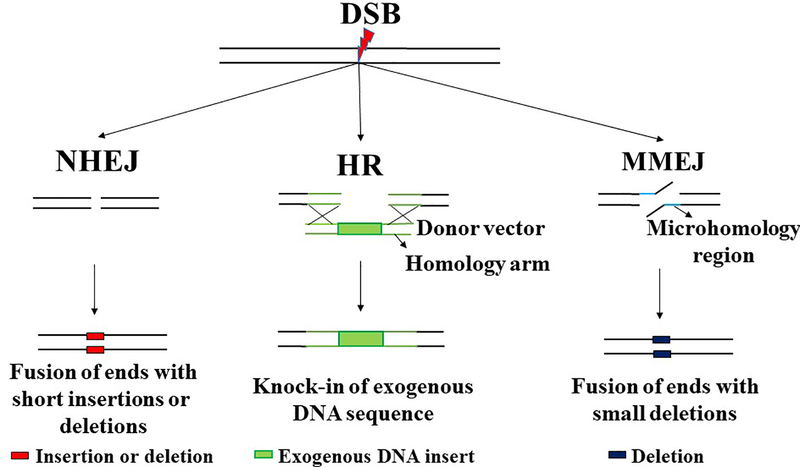
Figure 3: DNA repair mechanisms utilized for genome editing.
(A) NHEJ (Non-Homologous End Joining) is a double stranded break repair (DSB) pathway which causes fusion of the two ends directly without requiring a template for repair. NHEJ could be error prone as it usually creates short insertions or deletions (in red). For genome editing, NHEJ is used to create large deletions (2 DSBs) or short deletions (1 DSB) usually for correcting frameshift mutations and thereby restoring the reading frame in a subset of targeted cells. (B) HR (Homologous recombination) is a DSB repair pathway that utilizes a homologous template to exchange sequence information and is thus usually error free. For genome editing, HR is used for knocking in an exogenous DNA sequence by providing exogenous donor vector with the insert (green box) flanked by homology arms (green line). Single stranded oligonucleotides can also be used as a template for HR. (C) MMEJ (Microhomology Mediated End Joining) is an error prone DSB repair pathway that utilizes the microhomology region (2–25 bp) to repair and thereby cause deletion of the flanking sequence. For genome editing, MMEJ can be utilized to create small deletions (blue box) for correcting frame shift mutations caused by microduplication. Alternatively, MMEJ can also be used for knock-in of exogenous DNA sequence.