Figure 3. Differential stimulation of CD8+ T cells with distinct specificity of the functional peptides.
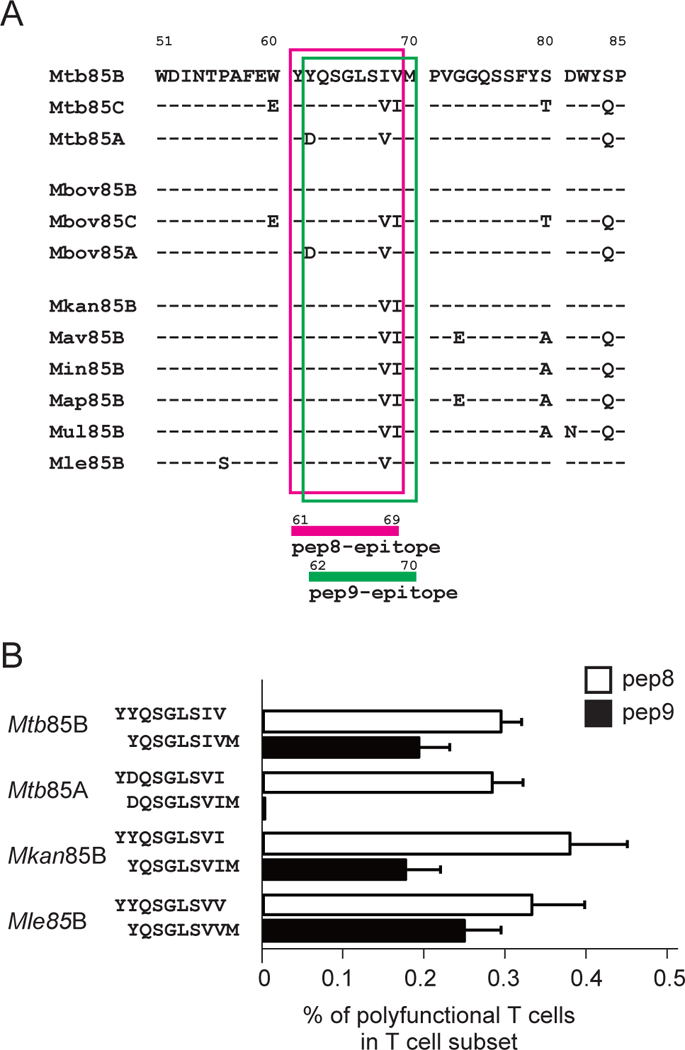
(A) Alignment of the epitope pep8 and pep9 for polyfunctional CD8+ T cell-induction on mycobacterium Ag85B complex. Amino acid sequence alignment of Mtb Ag85B (Mtb85B), Mtb Ag85C (Mtb85C), Mtb Ag85A (Mtb85A), M. bovis Ag85B (Mbov85B), M. bovis Ag85C (Mbov85C), M. bovis Ag85A (Mbov85A), M. kansasii Ag85B (Mkan85B), M. avium Ag85B (Mav85B), M. intracellulaire Ag85B (Min85B), M. avium subsp. paratuberculosis Ag85B (Map85B), M. ulcerans Ag85B (Mul85B) and M. leprae Ag85B (Mle85B). Amino acid sequence of Mtb85B and its differences from other mycobacterium sequences are shown in single letter code. pep8 and pep9 epitopes of mycobacterium Ag85B complex are highlighted with magenta and green line-rectangles, respectively. (B) Cross-reactivity and functionality between pep8 and pep9 for the induction of polyfunctional CD8+ T cells in immunized animals. The second position of the aspartic acid (D) at amino acid 62 in antigen 85A complex is critical for activation of CD8+ T cells in H2d mice, whereas those of the tyrosine (Y) at amino acid 62 in Ag85B complexes of Mtb85B, Mkan85B and Mle85B are cross-reactive. Substitutions of the eighth and ninth positions of valine (V) isoleucine (I) with either VI, IV and VV functions as epitope peptides. Data represent three independent experiments with five mice per group. Error bars represent SEM.
