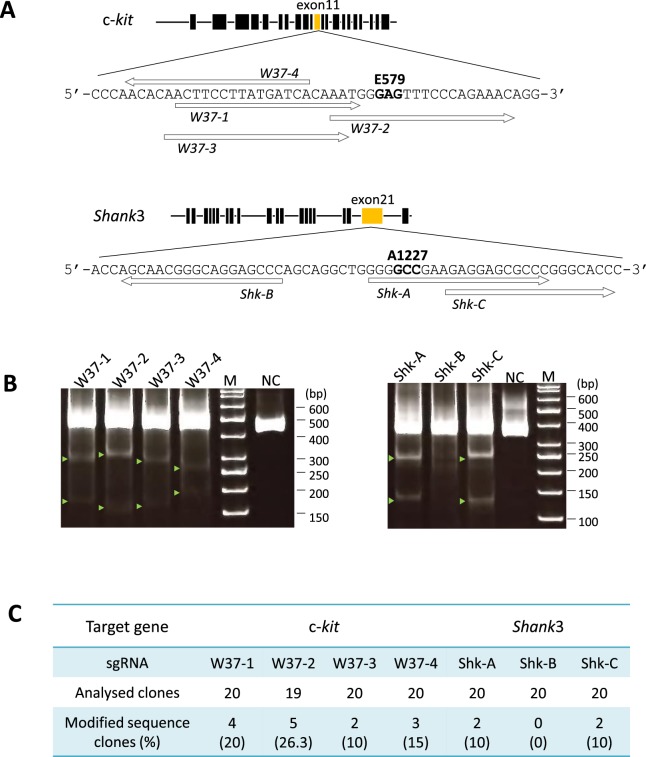
Figure 1.
Validation of each sgRNA sequence using marmoset fibroblast cells. (A) Schema of linearized target genes including exons (black boxes) and single-guide RNA (sgRNA) positions (white arrows). The marmoset c-kit gene (top) is located on chromosome 3 and contains 21 exons. 579 glutamic acid (E579, GAG) is located on exon 11 (yellow box). Marmoset Shank3 is located on chromosome 1 and contains 22 exons and 1227 alanine (A1227, GCC) is located on exon 21 (yellow box). (B) Cleavage activities of each sgRNA in marmoset fibroblast cells. The target gene modification by each sgRNA with hCas9 was examined using marmoset fibroblast cells and CEL-1 assay to confirm cleavage activity of designed sgRNAs. Green arrowheads indicate shifted bands compared to negative control, indicating target gene modification by CEL-1 assay. M; size marker, NC; negative control – PCR product obtained using wild-type marmoset tissue as a PCR template. (C) Rate of modified sequence clones in each target gene by subcloning and sequence analysis using PCR products of marmoset fibroblast cells.