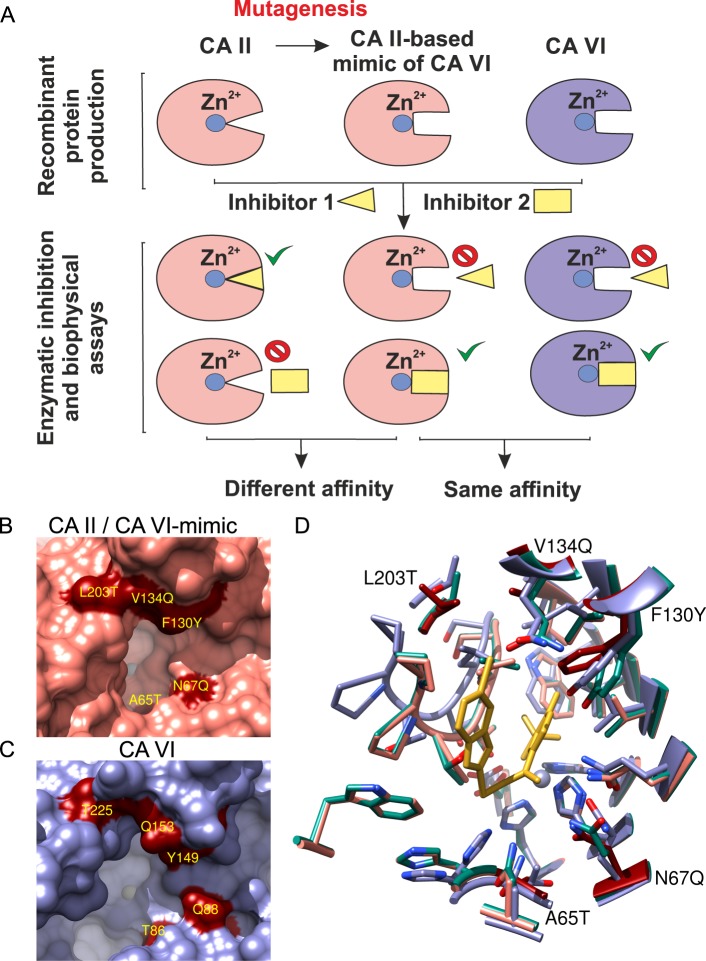
Figure 1.
(A) The mimic of CA VI was prepared from CA II by site-directed mutagenesis of amino acids that differ between two CA isoforms. The CA VI-mimic protein served as a model of compound binding to CA VI. (B) Active site of CA II (PDB ID: 3KS3). Dark red molecular surfaces mark the positions of point mutations introduced in the active site of CA II to resemble CA VI by making a multiple-residue mutant of CA II (CA VI-mimic). (C) Active site of CA VI (PDB ID: 3FE4). The light blue areas are buried molecular surfaces between interacting molecules in the homodimeric complex. Dark red molecular surfaces mark the equivalent positions between multiple-residue mutant of CA II (CA VI-mimic) and CA VI. The labels belong to CA VI (CA II numbering). (D) Superposed structures of the binding pockets of CA II (rose; PDB ID: 3M96), CA VI (blue; PDB ID: 3FE4), and CA VI-mimic (green; PDB ID: 6QL2). The mutated residues of CA II are colored dark red. The zinc ion in the active site of each CA isoform is shown as a grayish sphere in panels (B–D).