Fig. 1.
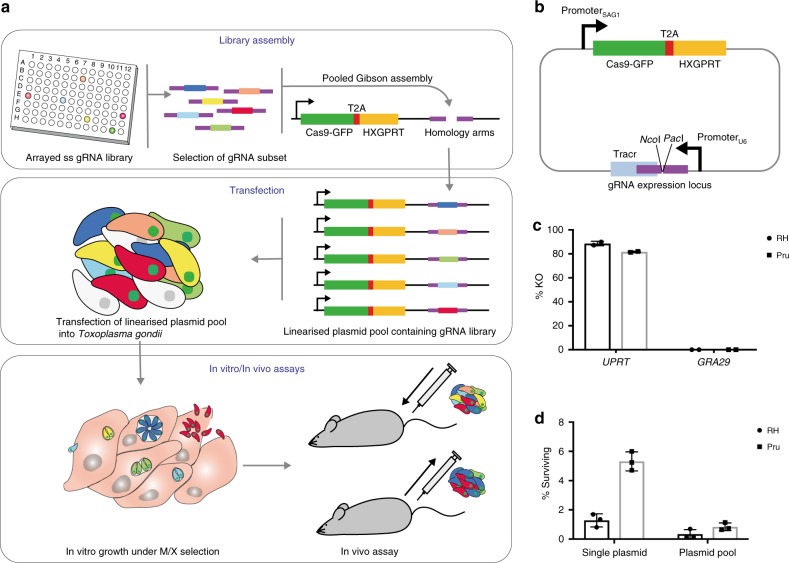
Single-step CRISPR knockout pools from arrayed single-stranded oligos. a Schematic of tailored CRISPR screening method. gRNA encoding oligonucleotides are arrayed in multiwell plates and the gRNA of interest are selected for pooled Gibson cloning into pCAS9-T2A-HXGPRT. The vector library is transfected into parasites to generate a pool of mutants for testing in vitro and in vivo. b Schematic of pCAS9-T2A-HXGPRT. Regions of homology used for Gibson cloning of oligonucleotides are shown in purple. c Gene disruption using integrated pCas9-T2A-HXGPRT. RHΔhxgprt and PruΔhxgprt were transfected with vectors targeting UPRT or a control gene, GRA29. Transfectants were grown under M/X selection for 6 days before performing a plaque assay under 5-fluorodeoxyuridine (FUDR) selection to test for UPRT disruption. The percentage of UPRT knockout (KO) was calculated by comparing plaques numbers in the presence and absence of FUDR. The mean is shown with SD error bars. n = 2 biologically independent experiments. d Parasite survival is reduced after transfection of plasmid pools. RHΔhxgprt and PruΔhxgprt were transfected with 10 μg pCAS9-T2A-HXGPRT targeting UPRT or a mixed pool of 1000 gRNA. Parasites were grown in the presence of M/X and the resultant plaques counted after 7 days. The percentage of parasites surviving compared to parasites seeded was calculated in a plaque assay. The mean is shown with SD error bars. n = 3 biologically independent experiments. See also Supplementary Fig. 2. Source data are provided as a Source Data file