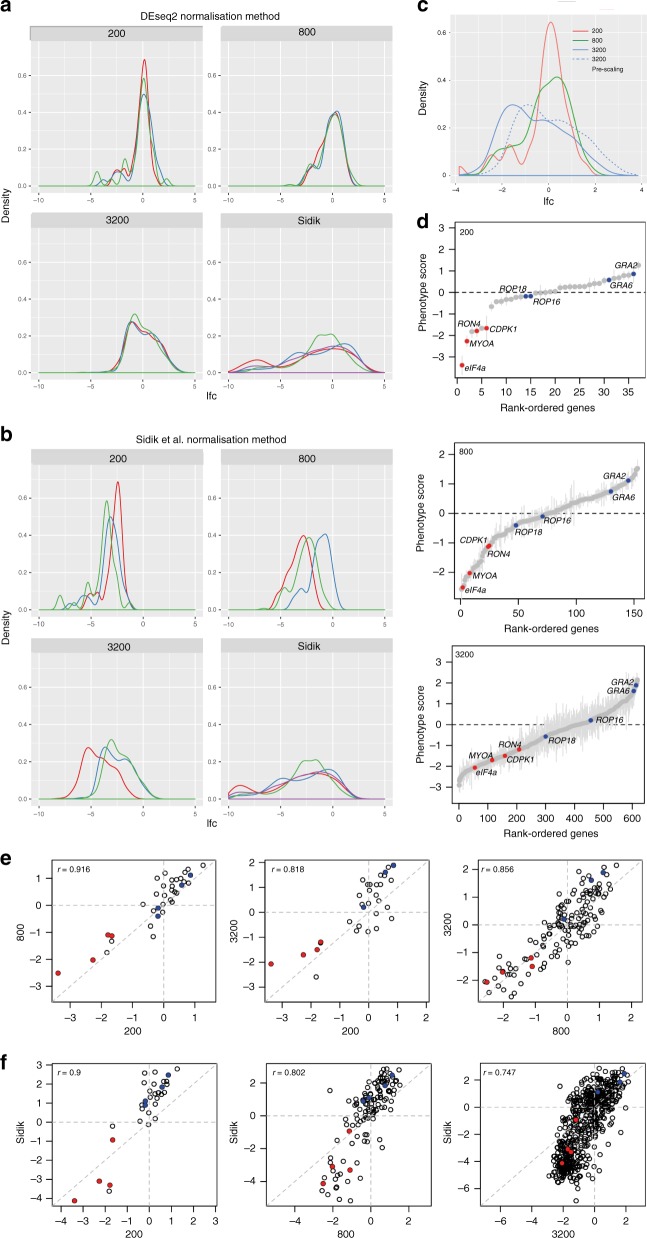
Fig. 2.
Variable pool sizes produce reproducible CRISPR selection results. PruΔhxgprt were transfected with pCAS9-T2A-HXGPRT carrying 200, 800 or 3200 gRNA in triplicate transfections. The log 2 fold change (lfc) of the gRNA presence after 6 days growth in M/X compared to the DNA input is shown. Normalisation of sequencing counts using either a DESeq2 or b total read count for the 200, 800 and 3200 libraries and the published Sidik et al.8 dataset. Density plots show improved reproducibility between replicates for small libraries using DESeq2. c Overlay of 200, 800 and 3200 library gene lfcs with (solid line) and without (dotted line) centering of the 3200 library. d Ranked-order plots showing gene lfcs for 200, 800 and 3200 gRNA libraries. Essential and non-essential control genes are marked in red and blue, respectively. Error bars represent SEM. n = 3 technical replicates. e, f Correlation plots with Pearson’s correlation coefficient, r, of gene lfcs show that phenotypes are reproducible e across library sizes and f correlate well with published phenotype scores. Essential and non-essential control genes are marked in red and blue, respectively. See also Supplementary Fig. 3 and Source data in Supplementary Data 2