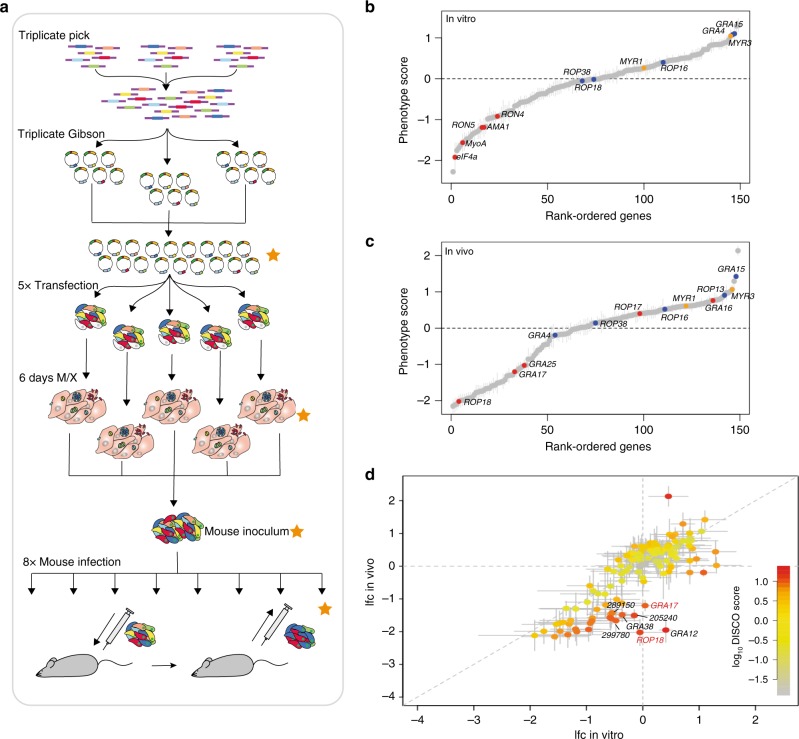
Fig. 3.
In vivo CRISPR screen identifies known and novel virulence factors. a Schematic of experimental design. gRNA were picked in triplicate, combined and used in triplicate Gibson cloning reactions. Vector pools were combined and transfected into PruΔhxgprt in five replicates. After 6 days under M/X selection, subsets were taken for sequencing and remaining parasites were combined to generate the mouse inoculum. Eight mice were infected intraperitoneally (i.p.) with 200,000 parasites and the parasites retrieved from the peritoneum after 5 days. Orange stars indicate samples analysed by sequencing. b, c Rank-ordered plot of gene lfcs in vitro (b) and in vivo (c). Essential and non-essential control genes for in vitro and in vivo growth marked in red and blue, respectively. Translocon components MYR1 and MYR3 marked in orange. Error bars represent SEM. n = 5 technical replicates. d Discordance plot showing gene lfcs in vitro and in vivo. Colour indicates strength of discordance score. Error bars show 95% confidence range. Examples of known and newly identified virulence factors labelled with red and black text, respectively. See also Supplementary Fig. 4 and Source data in Supplementary Data 3