Fig. 4.
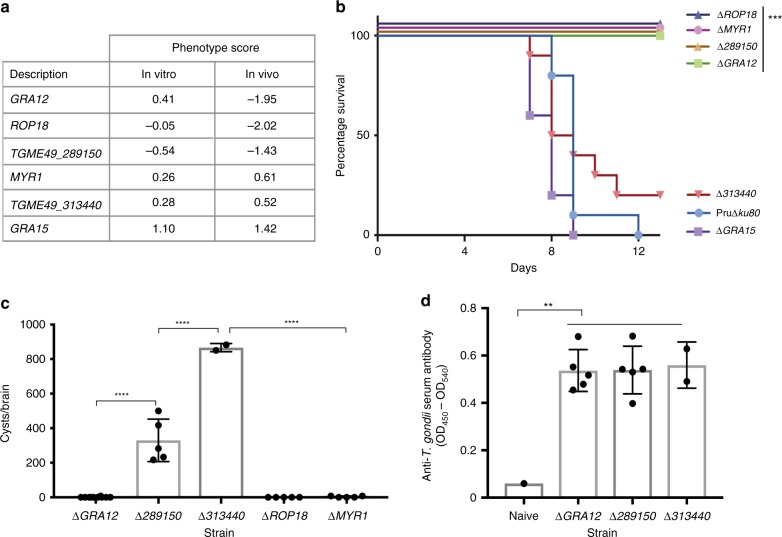
Confirmation of virulence defects identified in the CRISPR pool in vivo. a In vitro and in vivo phenotype scores for genes targeted for knockout (KO) and mouse infection. b C57BL/6 mice were infected intraperitoneally (i.p.) with 50,000 tachyzoites of the parental line PruΔku80 (two experiments; n = 10 mice) and ΔGRA12 (two experiments; n = 10 mice), ΔTGME49_289150 (one experiment; n = 5 mice), ΔTGME49_313400 (two experiments; n = 10 mice) and control strains ΔGRA15 (one experiment; n = 5 mice), ΔROP18 (one experiment; n = 5 mice) and ΔMYR1 (one experiment; n = 5 mice). ΔROP18, ΔMYR1, ΔTGME49_289150 and ΔGRA12 were significantly different from PruΔku80 with P values of 0.0075, <0.0001, 0.0004 and <0.0001, respectively, using the Mantel–Cox test. c Brains were isolated from surviving mice and cysts enumerated at 24 or 28 days post infection. ΔTGME49_313400 n = 2, ΔGRA12 n = 10, ΔTGME49_289150 n = 5. P values were <0.0001 as calculated by one-way analysis of variance (ANOVA) with Tukey’s multiple comparison test. d Serum was isolated from mice surviving acute infection and tested for antibodies against Toxoplasma-soluble antigens in an ELISA. P values were ΔGRA12 P = 0.006, ΔTGME49_289150 P = 0.0058 and ΔTGME49_313400 P = 0.0009 compared to the naive sample as calculated by one-way ANOVA with Tukey’s multiple comparison test. ** refers to P < 0.01, *** to P < 0.001 and **** to P < 0.0001. See also Supplementary Figs. 5 and 6. Source data are provided as a Source Data file