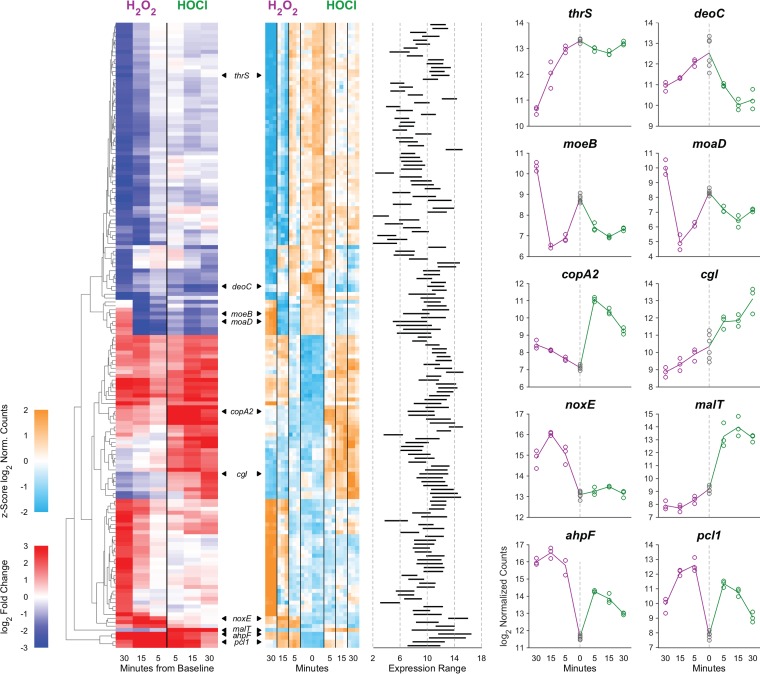
FIG 5.
Cluster analysis of gene expression levels in anaerobically grown L. reuteri ATCC PTA 6475 treated with 0.12 mM H2O2 (purple) or 1.25 mM HOCl (green) 0, 5, 15, and 30 min after stress treatment (times plotted outward from the center for each panel). A total of 160 genes are shown that had a 4-fold change in expression and Bonferroni-corrected P value of <0.01 for at least one of the six treatment time point comparisons against baseline. The red-blue heat map at the left shows the fold changes (color) of these 160 genes (rows) for each of the six pairwise comparisons (columns). Clustering was performed to illustrate the diversity of response profiles rather than to establish a number of “canonical” patterns. Genes (rows) were hierarchically clustered based on Euclidean distance and average linkage. The second, teal-orange heat map shows the per-gene z-score (the number of SDs away from the mean of expression in the reference) of log2 normalized data of genes (in the same order) for each replicate of each treatment/time point and baseline (columns). The next plot shows the range of the log2 normalized expression data (of averaged replicates) for each of the genes. Expression data for 10 representative genes are plotted to the right (symbols, individual replicates; lines connect means of replicates). Called-out genes are thrS (LAR_RS09850), deoC (LAR_RS00565), moeB (LAR_RS05335), moaD (LAR_RS05395), copA2 (LAR_RS02280), cgl (LAR_RS01550), noxE (LAR_RS00345), malT (LAR_RS00275), ahpF (LAR_RS05795), and pcl1 (LAR_RS08080).