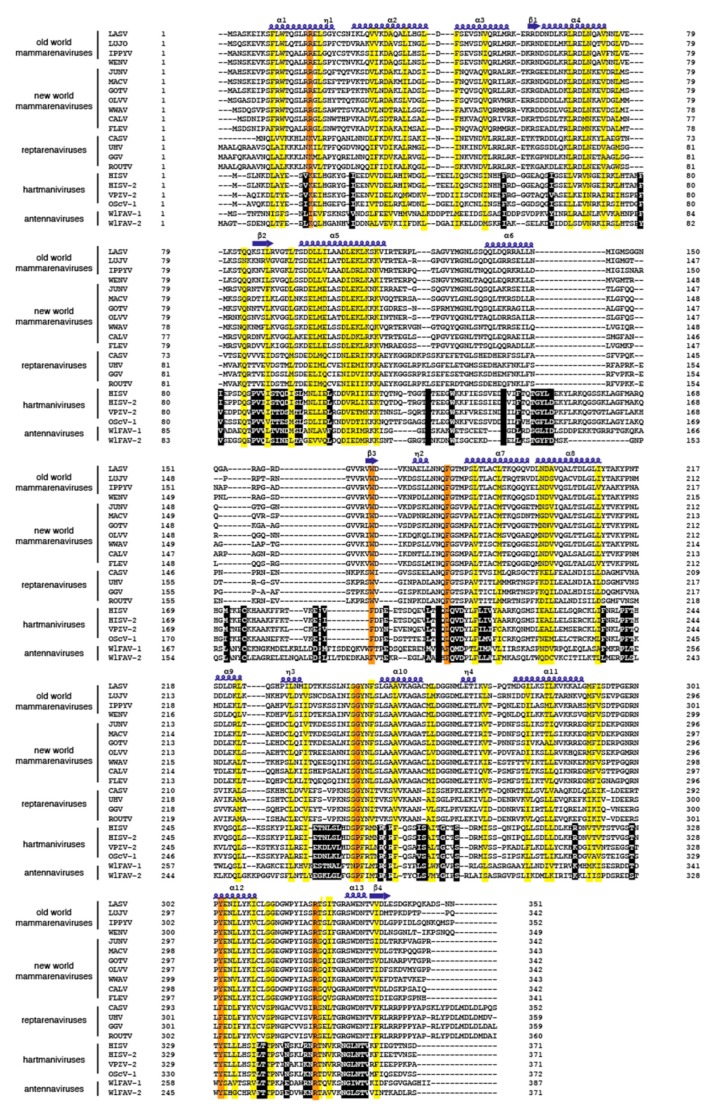
Figure 5.
Amino acid sequence alignment of the N-termini of nucleoproteins from members of the four arenavirus genera. Virus abbreviations: Lassa, LASV; Lujo, LUJV; Ippy, IPPYV; Wenzhou, WENV; Junin, JUNV; Machupo, MACV; Guanarito, GOTV; Olivaros, OLLV; Whitewater Arroyo, WWAV, California (Pichinde), CALV; University of Helsinki, UHV; Golden Gate, GGV; Haartman Institute snake, HISV and HISV-2; Old Schoolhouse, OScV; Wēnlǐng Frogfish arenavirus, WlFAV-1 and WlFAV-2. Residues that are conserved (identical or chemically similar) are highlighted in yellow. Conserved residues involved in RNA binding are highlighted in orange. Blue springs represent helical structures and blue arrows are beta sheets. Similar residue/sequences in antennaviruses and hartmaniviruses not present in NP of members from other genera are highlighted in black.