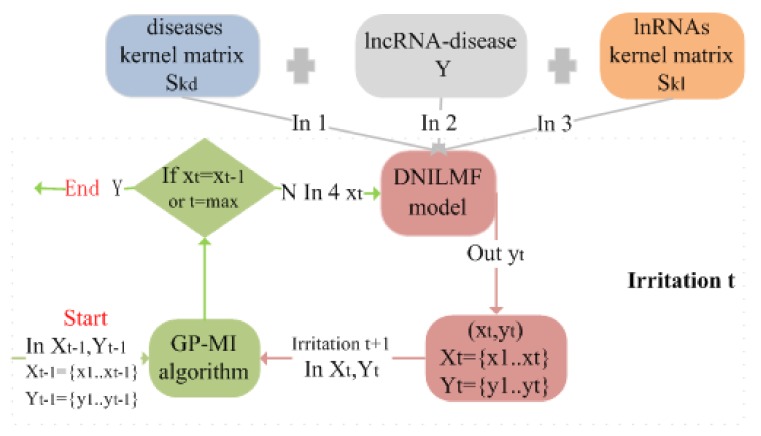
Figure 2.
The optimization process of GP-MI for the DNILMF model parameters. At irritation t: Step 1: get according to the previous query and observations ; Step 2: if = or t is equal to the max value, exit the program; if not, put , the disease kernel matrix, lncRNA–disease association matrix, and lncRNA kernel matrix into the DNILMF model, and we can get output . Then, take and as the start of the next irritation.