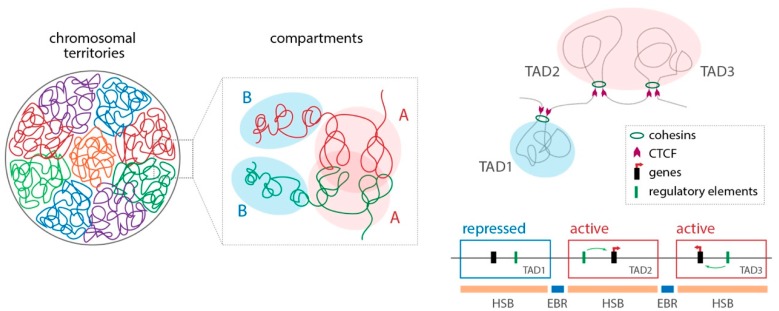
Figure 3.
The integrative breakage model, a multilayer framework for the study of genome evolution that takes into account the high-level structural organisation of genomes and the functional constraints that accompany genome reshuffling [64]. Genomes are compartmentalised into different levels of organisation that include: (i) chromosomal territories, (ii) ‘open’ (termed ‘A’)/’closed’ (termed ‘B’) compartments inside chromosomal territories, (iii) topologically associated domains (TADs) and (iv) looping interactions. TADs, which are delimited by insulating factors such as CTCF and cohesins, harbour looping topologies that permit long-range interactions between target genes and their distal enhancers, thus providing ‘regulatory neighbourhoods’ within homologous syntenic blocks (HSBs). In this context, the integrative breakage model proposes that genomic regions involved in evolutionary reshuffling (evolutionary breakpoint regions, EBRs) which will likely be fixed within populations are (i) those that contain open chromatin DNA configurations and epigenetic features that could promote DNA accessibility and therefore genomic instability, and (ii) that do not disturb essential genes and/or gene expression.