Abstract
The pleurocarpous feather moss Pleurozium schreberi is a ubiquitous moss species which plays a fundamental role in many terrestrial ecosystems, for instance within the boreal forest, the Earth’s largest terrestrial biome, this species plays a significant role in driving ecosystem nitrogen and carbon inputs and fluxes. By hosting dinitrogen (N2)-fixing cyanobacteria, the moss-cyanobacteria symbiosis constitutes the main nitrogen input into the ecosystem and by the high productivity and the low decomposability of the moss litter, P. schreberi contributes significantly to build-up soil organic matter, and therefore long-term C sequestration. Knowledge on P. schreberi genome will facilitate the development of ‘omics’ and system’s biology approaches to gain a more complete understanding of the physiology and ecological adaptation of the moss and the mechanisms underpinning the establishment of the symbiosis. Here we present the de novo assembly and annotation of P. schreberi genome that will help investigating these questions. The sequencing was performed using the HiSeq X platform with Illumina paired-end and mate-pair libraries prepared with CTAB extracted DNA. In total, the assembled genome was approximately 318 Mb, while repetitive elements account for 28.42% of the genome and 15,992 protein-coding genes were predicted from the genome, of which 84.23% have been functionally annotated. We anticipate that the genomic data generated will constitute a significant resource to study ecological and evolutionary genomics of P. schreberi, and will be valuable for evo-devo investigations as well as our understanding of the evolution of land plants by providing the genome of a pleurocarpous moss.
Keywords: Pleurozium schreberi, Genome sequencing, Comparative genomic, Annotation, Genome assembly
Pleurozium schreberi (Brid.) Mitt. also known as the red-stemmed feather moss belongs to the order Hypnales, family Hylocomiaceae. P. schreberi has a wide global distribution and is one of the most common moss species dominating the ground layer in the boreal forest, sub-alpine and Arctic ecosystems (Nilsson and Wardle 2005; Lindo and Gonzalez 2010). P. schreberi serves a variety of key functions in its ecosystem; for instance, P. schreberi can host dinitrogen (N2)-fixing cyanobacteria, a symbiosis which serve as the major input of nitrogen (N) into boreal forests and thus play a vital role in primary productivity of this N limited ecosystem (Turetsky 2003; Lindo et al. 2013). High-resolution secondary ion mass spectrometry verified transfer of fixed N from the cyanobacteria to the moss host, further demonstrating that the symbiosis governs the main N entrance to boreal forests (Bay et al. 2013). While, the genetic and genomic of P. schreberi symbiotic cyanobacteria have been previously studied (Ininbergs et al. 2011; Warshan et al. 2016, 2017 and 2018), nothing is known about the moss genomic diversity and gene repertoire needed to form the symbiosis. In particular the genome will be a crucial tool for further studies on the symbiotic interaction with N2-fixing cyanobacteria where previous study, have shown the importance of moss secreted signaling molecules to attract the cyanobacteria (Bay et al. 2013). Further, the biomass of P. schreberi and Hylocomium splendens (another ubiquitous feather moss in the boreal forest) can account for up to a third of the total forest productivity (Goodale et al. 2002). As boreal forests represent one of the largest terrestrial carbon (C) sinks on Earth, storing approximately 30% of total terrestrial C stocks (Goodale et al. 2002; Pan et al. 2013) the feather mosses play a significant role in the global C cycles. As such, the response of the feather moss cover to future climate change may influence whether and how the ecosystem will be regulated and the consequences for C sequestration by this biome (Lindo et al. 2013). The availability of P. schreberi genomic resource will facilitate population genetic study of P. schreberi at genomic level using genotyping-by-sequencing approaches. For instance, investigating the link between intraspecific genetic diversity and moss traits variations, such as its physiology, decomposition rate and C accumulation could reveal pattern of local adaptations that might have an impact on N and C cycles. Even though mosses are the second most diverse phylum of land plant with approximately 13,000 species (Goffinet et al. 2009), to date only two other moss species had their genome sequenced i.e., Sphagnum fallax (DOE-JGI, http://phytozome.jgi.doe.gov/; Shaw et al. 2016), and Physcomitrella patens (Rensing et al. 2008). The latter of which have become a very important model organism (Cove et al. 2009).
In this study, we present a draft genome of an axenic pleurocarpous feather moss P. schreberi. Indeed, the sequenced draft genome of the moss will be a fundamental resource for future research spanning from evolutionary to ecological aspects.
Materials and Methods
Sample collection
Pleurozium schreberi is a pleurocarpous moss with typical tissue growth such as leafy gametophores with phyllids, rhizoids and reproductive organs (Figure 1A, B), protonemal tissue (Figure 1C) as well as gametangia (Figure 1D). Intact P. schreberi sporophytes were randomly collected June 26, 2016 on Blidö (59°62’20.06”N, 18°90’26.41”E), an island located in the Stockholm archipelago, Sweden. An axenic line was generated by first separating the septa from the sporophyte with forceps and then placing in 99.9% ethanol for 1 min, subsequently moved to 1 min in 5% sodium hypochlorite followed by 4 rounds of rinsing in sterile water. The sterilized spores where placed on BCD media (Cove et al. 2009) + 5% sucrose for approximately 2 weeks. Axenic germinated spores were transferred to BCD and placed in an incubator at 24° with constant white light at 30 W/m2 to allow the gametophore to growth. Pictures of the P. schreberi tissues were taken using the Zeiss Stemi 2000-C Stereo microscope equipped with an AxioCam HR microscope camera (Zeiss, Oberkochen, Germany).
Figure 1.
Different growth morphology of P. schreberi. (A) Wild type P. schreberi. (B) Axenic P. schreberi growing on BCD media showing the gametophore, rhizoids, phyllids and reproductive bundles. (C) Protonemal tissue and (D) male gametangia.
Genome sequencing and assembly
DNA extraction was performed from gametophores originating from a single spore isolate according to Schlink and Reski (2002) with the following modifications; 10% CTAB was used during the extraction and DNA was re-dissolved in water. After the DNA quality check using the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, USA), 180bp average fragment size Illumina TruSeq PCR-free library (Illumina, Cambridge, UK), 3kb and 5-8kb insert size Nextera mate-pair libraries (Illumina) were constructed at SciLifeLab (Stockholm, Sweden) following manufacturer’s recommendations. Sequencing proceeded on the Illumina HiSeq X platform, where the Illumina TruSeq PCR-free library and the Nextera mate-pair libraries were sequenced on separate lanes at 2x150bp. Preliminaries de novo assemblies were conducted with NouGAT, NGI open universal Genome Assembly Toolbox pipeline (https://github.com/vezzi/NouGAT) using Abyss (Simpson et al. 2009), Allpaths (Butler et al. 2008), and SOAPdenovo (Luo et al. 2012) assemblers with default settings. The quality of the selected assembly was assessed using BUSCO version 3 (Simão et al. 2015) with the eukaryota database (release 9) in protein mode. We estimated genome size using k-mer counting of quality and barcode trimmed reads using Jellyfish v2.2.10 (Marçais and Kingsford 2011). K-mer frequency distributions of 21-, 25-, 31-, 41-, 65-mers were generated, then we used findGSE (Sun et al. 2017) to estimate genome size and repeat content.
Repeat content analysis
To identify repeats in the genome assembly, a custom species-specific repeat library was created, using the RepeatModeler package 1.0.8 (Smit 2010) with default parameters. As repeats can be part of actual protein-coding genes, the candidate repeats were vetted against the Uniprot/Swiss-prot proteins set (minus transposons) to exclude any nucleotide motif stemming from low-complexity coding sequences. From the repeat library, identification of repeat sequences present among the genome was performed using RepeatMasker 4.0.3 (Smit et al. 2010) and RepeatRunner (Magrane and Consortium 2011).
Gene prediction
The gene models were carried out using the MAKER pipeline, version 3.01.1 (Cantarel et al. 2008; Holt and Yandell 2011). The gene models incorporate ab initio gene prediction, homology-based prediction and RNA-seq assisted prediction. Prior to ab initio gene prediction, repeat regions of the moss were masked based on repeat annotation results. Homology-based prediction and RNA-seq assisted prediction used proteins sequences collected on the Uniprot database (Magrane and Consortium 2011) selecting those belonging to the Swiss-prot section and nine P. schreberi transcriptomes coming from the JGI study ID number Gs0110198. The transcriptome of P. schreberi (biosample ID Gb0144502- Gb0144504) had already been assembled by JGI using Rnnotator 3.4.0 (Martin et al. 2010) and completed with Velvet 1.2.07 (Zerbino and Birney 2008). The six other transcriptomes (ID Gb0144505- Gb0144510) have been assembled using HISAT2 (version 2.1.0) (Kim et al. 2015) and StringTie 1.2.2 (Pertea et al. 2015) with default parameters. MAKER ab initio training was performed using Augustus (Stanke et al. 2008), GeneMark (Besemer and Borodovsky 2005) and SNAP (Bromberg and Rost 2007) with transcripts and high-confidence proteins data. For ab initio training purpose, a gene set was created by selecting the best gene models based on i) genes have to be complete (i.e., start/stop codons mandatory); ii) no similarity over 85 is allowed among genes of the set; iii) Annotation Edit Distance (AED) scores have to be inferior to 0.3; iv) genes have to be at a distance of 1,000 bp from each other. In total, 4,264 genes were selected and used for Augustus and SNAP training process. GeneMark has also been trained using splicing sites information of nine transcriptome assemblies. Then, an evidence-guided build was computed by allowing the MAKER software to construct gene models directly from both aligned transcript sequences and reference proteins. Finally, a second round of annotation using MAKER with ab initio tools Augustus, SNAP, GeneMark and EvidenceModeler (Haas et al. 2008) and evidence build (proteins and transcripts) allowed creating an ab initio evidence-driven gene build. After a visual analysis of the ab initio evidence-driven gene build, spurious ORFs among the gene structure were discovered in some loci, and fixed using in-house scripts (https://github.com/NBISweden/GAAS). In the case of loci containing proteins and/or transcripts without ab initio prediction, the gene models were taken from the evidence build.
Gene annotation
The functional inference for genes and transcripts were performed using the translated CDS features of each coding transcript. Each predicted protein sequences were blasted against the Uniprot/Swissprot database to retrieve the gene name and the protein function as well as run against InterProscan 5.21-60 (Jones et al. 2014) to retrieve functional information from 21 different sources (Table S1).
Gene name inference was performed with the best blast hit approach using the Uniprot reference data set (Kim et al. 2015). Only the hits with an e-value inferior to 10E-7 were taken into account. tRNAs have been predicted by using tRNAscan v1.3.1 (Lowe and Eddy 1997) with default settings and other non-coding RNAs by using the RNA family database Rfam (Nawrocki et al. 2015) with conserved eukaryotic models only.
Analyses of potential whole genome duplication in P. schreberi
Whole genome duplication (WGD) events can be detected using synonymous substitution rates (Ks) among pairs of paralogous genes. WGD event(s) within P. schreberi were estimated using the FASTKs pipeline (https://github.com/mrmckain/FASTKs). Protein-coding genes were blasted against themselves using an e-value cutoff of 10E-5 and then putative pairs were filtered using FASTKs default parameters. Amino acid sequences for putative paralog pairs were then aligned using MUSCLE v3.8.31 (Edgar 2004), and back translated to CDS using PAL2NAL v14 (Suyama et al. 2006). Ks were estimated for the aligned pairs using codeml in PAML v4.8 (Yang 2007) using the same parameters used by McKain et al. (2012). All Ks values ≤0.1 were excluded for analysis to avoid the incorporation of allelic variants and to prevent the fitting of a component to infinity, while Ks values >5.0 were removed because of Ks saturation (Vanneste et al. 2015). Normal mixture models were estimated for Ks values using the mclust v.5.4.3 (Scrucca et al. 2016) in R (v3.5.1). We evaluated mixture models with between one and nine components, and the best fit model was chosen using the Bayesian Information Criterion (BIC).
Comparative genomic
A comparative genomic analysis was performed on three different moss genomes (Physcomitrella patens, Sphagnum fallax and P. schreberi) using OrthoMCL 2.0.9 (Li et al. 2003) with mcl-14-137 (Enright et al. 2002) and BLAST+ 2.2.28 (Camacho et al. 2009) to infer homologous (both orthologous and paralogous) relationships among a set of protein sequences. The input data used included protein sequences from P. schreberi, P. patens (v1.39) and S. fallax (v0.5). For each moss genome, the CDS were filtered to keep only the longest coding sequence for each gene, and the corresponding sequences were extracted, translated and filtered to retain those larger than 50 amino acids (Table S2). An all-against-all BLASTP analysis of the sequences of these moss species led to the identification of candidate homologs. BLAST hits with an e-value >10E-5 and for which the query and the hit sequence had <50% overlap of their gene length, were excluded. Gene families characterized by the OrthoMCL analysis were processed to infer the presence/absence of the genes along a species tree to provide an evolutionary view of the gene flux in those genomes.
Data availability
The raw data are deposited in NCBI with SRA accessions numbers; SRR8297981 and SRR8297982 (BioProject accession number PRJNA509035). The BioSample is available with accession number SAMN10578977 at NCBI. The assembled genome is available with the accession number VACF00000000 at NCBI. All of the annotation files are available at github: https://github.com/PycnopodiaD/Pleurozium_schreberi_annotated_genome_files. Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.7775951.
Results and Discussion
Genome assembly
Concerning the initial genome assemblies of Pleurozium schreberi, the Allpaths assembler showed the best assembly statistics (i.e., highest NG50 value and lowest number of scaffolds) compared to assemblies done by Abyss and SOAPdenovo (Table S3). Quality control of the Allpaths assembly including coverage distribution, GC content, contig length and median coverage are shown in Figure S1. Low coverage contigs showed deviation in GC content, which might be a sign of contamination, and therefore they were removed from the assembly (Figure S1C). The total length of the assembly was 318.34 Mb in 2,695 scaffolds with an NG50 score of 204 Kb, and a genomic GC content of 26.4% (Table 1). The repeat regions were estimated to account for 28.42% of the genome (90 Mb; Table 2), and BUSCO analysis captured 90.1% of genes in the eukaryota database with 75.6% complete, 14.5% partial and 9.9% missing genes (Table 3). Even if the genome size and number of repetitive elements in P. schreberi appears smaller compared to P. patens, they are similar to the estimates of S. fallax assembly (genome size of 475.8 and 396 Mb, and percentage of repetitive elements of 57% and 32% for P. patens and S. fallax, respectively; Shaw et al. 2016; Lang et al. 2018). When comparing to the k-mer analysis, the predicted genome size and percentage of repetitive elements are reasonably close to our assembly (Table S4). Genome size estimates ranged from 296.9 to 319.4 Mb, and repetitive elements ranged from 12 to 26% of the genome (Table S4). Nevertheless, the length of our assembly is small compared to genome size predicted by flow cytometry that indicated a genome 1.3x – 2.5x larger (ranging from 418 – 809 Mb; Voglmayr 2000; Bainard 2011). A high proportion of repeat sequences in P. schreberi genome could result in an underestimation of genome size, but k-mer analysis provides an indication that this was not the case, since the percentage of predicted repeated elements was very similar to the one estimated by our repeat analysis (Table 2 and Table S4). Alternatively, since the BUSCO analysis indicates that our genome assembly was incomplete, the discrepancy observed with the flow cytometry estimates could be partially explained by the fraction of coding sequence missing from our data. Besides, considering that the genome size estimations by flow cytometry originate from gametophyte cells (Voglmayr 2000; Bainard 2011), a most likely explanation to the extreme variation observed in genome sizes seems to be endopolyploidy in gametophyte tissues of P. schreberi. In addition, different isolates could have different ploidy levels, particularly in widespread species like P. schreberi. This appears to be a common phenomenon in this species and in mosses in general (Bainard and Newmaster 2010). Throughout the assembly of a plant genome many difficulties are incurred (Griesmann et al. 2018) and it is therefore not surprising that this assembly is less complete at the first draft stage, though future work should be conducted to improve these assemblies.
Table 1. Statistics of Pleurozium schreberi genome assembly.
| Total length (bp) | 318,338,550 |
|---|---|
| Number of scaffolds | 2,695 |
| Number of uncalled bases (N’s) | 98,291,915 |
| Number of N-regions | 35,733 |
| NG50 (bp) | 204,181 |
| GC-content (%) | 26.4 |
Table 2. Repeat statistics by RepeatMasker and RepeatRunner.
| RepeatMasker | RepeatRunner | |
|---|---|---|
| Number of repeats | 255,686 | 484 |
| Total size (kb) | 90,223 | 271.39 |
| Mean size (bp) | 352.66 | 560.73 |
| Percentage of the genome (%) | 28.34 | 0.09 |
Table 3. Statistics of the genome completeness using BUSCO.
| Number of BUSCO groups | Percentage | |
|---|---|---|
| Total BUSCO groups searched | 303 | 100 |
| Complete BUSCOs | 229 | 75.6 |
| Complete Single-Copy BUSCOs | 167 | 55.1 |
| Complete Duplicated BUSCOs | 62 | 20.5 |
| Fragmented BUSCOs | 44 | 14.5 |
| Missing BUSCOs | 30 | 9.9 |
Genome annotation
The final annotation of the P. schreberi genome from the MAKER annotation pipeline included 15,992 protein-coding genes (Table 4). The functional annotation using InterProscan to detect motifs, domains, signatures, and BLASTP on the Uniprot/Swissprot database resulted in putative function annotation of 13,470 proteins (84.23% of the CDS; Table S5). Further, the best blast hit approach using the Uniprot/Swissprot database assigned a name to 12,295 genes (76.88%; Table S5).
Table 4. Coding genes annotation statistics.
| Description | |
|---|---|
| Number of protein-coding genes | 15,992 |
| Average CDS length (bp) | 1,015 |
| Average number of exons per mRNA | 5.7 |
| Average exon length (bp) | 220 |
| Percentage of the genome covered by: | |
| Gene | 15.90 |
| Exons | 6.30 |
| Introns | 9.30 |
Whole genome duplication in P. schreberi
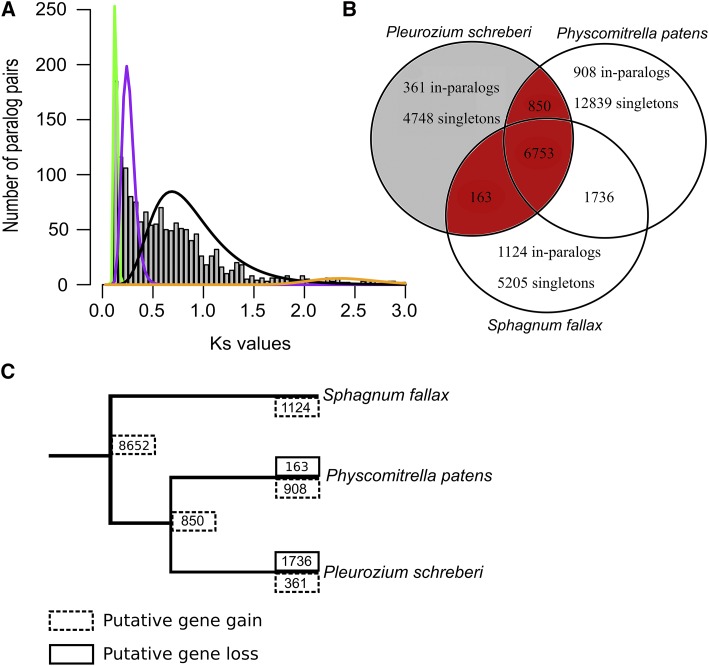
Our Ks-based analysis of paralogs was able to detect a peak of Ks values between 0.5 and 2.0, which support one WGD in P. schreberi (Figure 2A). Ks analyses conducted on P. schreberi and 24 other Hypnales transcriptomes, were unable to detect a clear evidence of WGD event (Johnson et al. 2016), but our analysis showed a stronger signal of WGD. When Gaussian distributions were fitted to the Ks values the best fit was of 4 components, but no obvious second large peak of Ks values was observed (Figure 2A). This would have been an indication of a second WGD event in P. schreberi, as observed in P. patens and S. fallax where several rounds of WGD and/or multiple large-scale duplication events have occurred (Devos et al. 2016; Lang et al. 2018). When considering that the number of protein-coding genes in the genome of P. schreberi is 2x smaller than in P. patens and 1.7x smaller than in S. fallax (32,926 and 26,939 CDS for P. patens and S.fallax, respectively), the fact that only one WGD event occurred in P. schreberi could explain the difference observed.
Figure 2.
Detection of whole genome duplication in P. schreberi, shared and unique orthologs, and gene flux between three moss genomes. (A) Distribution of synonymous substitution rates (Ks) among pairs of paralogous genes within P. schreberi. The solid curved lines are the inferred distributions from the mixture model, and colors represent the four component distributions under the model with the highest BIC score. (B) Venn diagram showing shared and unique gene families between moss species. Gene families shared between P. schreberi, P. patens and/or S. fallax are shown in red and the set unique to P. schreberi in gray. (C) Phylogenetic relationships between moss species and inferred gene families gain and loss in this lineage. The numbers of gain/loss events are represented by a solid and dotted box, respectively.
Comparative genome analysis
The comparison of gene content of the P. schreberi genome with the two other moss genomes available (P. patens and S. fallax) was conducted to illustrate potential gene flux between the moss species. We found 6,753 gene families shared between the moss species (Figure 2B, Table S2), and 5,109 gene families unique to P. schreberi (361 in-paralogs and 4,748 singletons). The comparative analysis suggests a large evolutionary difference between the three species, while Figure 2B shows that P. schreberi is sharing nearly 60% of gene families with P. patens and/or S. fallax, and that the remaining 40% of gene families identified in P. schreberi are unique for the species. Besides, it appears that more gene families are shared between S. fallax and P. patens (1,736 shared gene families) than with P. schreberi (Figure 2B). The gene flux analysis over the evolutionary time line for P. schreberi, S. fallax and P. patens, provides some indications to this observation by predicting that most of the gene gain occurred in the most recent common ancestor between the three moss species (8,652 gene families; Figure 2C), and that after the split with P. patens, P. schreberi gain 361 gene families and loss the 1,736 gene families that are in common between P. patens and S. fallax (Figure 2C). Nevertheless, this large putative gene loss in the genome might be due to the incompleteness of our draft assembly (Table 3). Moreover, due to the limited number of moss genomes available, this result remains highly speculative especially considering that in the moss phylogeny, P. schreberi, P. patens and S. fallax are evolutionarily quite distant from each other (Liu et al. 2019).
Conclusions
Illumina HiSeq X combining mate-pair and low error rate short-read sequencing enabled de novo assembly of the genome of a pleurocarpous moss species, Pleurozium schreberi.
Here, we provide a draft assembly that will be useful for long-read sequencing to improve genome scaffolding and completeness of P. schreberi genome. Further, a large part of the genes was functionally annotated (84.23%) and preliminaries gene ontology and comparative analyses were undertaken, which will ease future functional genomic studies. Considering that P. schreberi occupies a key role in the ecosystem functioning of cold biomes, the data generated provide the genomic resources to deepen our understanding of the physiology of this ecologically important moss species.
Acknowledgments
We would like to thank colleagues at the Oak Ridge National Laboratory, Duke University, and the DOE Joint Genome Institute for access to prepublication genome and annotation of Sphagnum fallax to use in comparisons against gene families. The Nilsson-Ehle Endowments from The Royal Physiographic Society of Lund for financial support. The authors would also like to acknowledge support from Science for Life Laboratory (SciLifeLab, Sweden), the National Genomics Infrastructure (NGI, Stockholm, Sweden), Nation Bioinformatics Infrastructure (NBIS; Stockholm, Sweden) and Uppmax (Uppsala, Sweden), for providing assistance in massive parallel sequencing as well as the computational infrastructure.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.7775951.
Communicating editor: S. Mathews
Literature Cited
- Bay G., Nahar N., Oubre M., Whitehouse M. J., Wardle D. A. et al. , 2013. Boreal feather mosses secrete chemical signals to gain nitrogen. New Phytol. 200: 54–60. 10.1111/nph.12403 [DOI] [PubMed] [Google Scholar]
- Bainard J. D., 2011. Patterns and biological implications of DNA content variation in land plants. Ph.D. Thesis, University of Guelph, Guelph, Ontario, Canada.
- Bainard J. D., and Newmaster S. G., 2010. Endopolyploidy in Bryophytes: Widespread in mosses and absent in liverworts. J. Bot. 2010: 7. [Google Scholar]
- Besemer J., and Borodovsky M., 2005. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 33: W451–W454. 10.1093/nar/gki487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y., and Rost B., 2007. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 35: 3823–3835. 10.1093/nar/gkm238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J., MacCallum I., Kleber M., Shlyakhter I. A., Belmonte M. K. et al. , 2008. ALLPATHS: De novo assembly of whole-genome shotgun microreads. Genome Res. 18: 810–820. 10.1101/gr.7337908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J. et al. , 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel B. L., Korf I., Robb S. M., Parra G., Ross E. et al. , 2008. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18: 188–196. 10.1101/gr.6743907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cove D. J., Perroud P.-F., Charron A. J., McDaniel S. F., Khandelwal A. et al. , 2009. The moss Physcomitrella patens: A novel model system for plant development and genomic studies. Cold Spring Harbor Protoc. 2009: pdb.emo115. [DOI] [PubMed]
- Devos N., Szövényi P., Weston D. J., Rothfels C. J., Johnson M. G. et al. , 2016. Analyses of transcriptome sequences reveal multiple ancient large-scale duplication events in the ancestor of Sphagnopsida (Bryophyta). New Phyt. 211: 300–318. [DOI] [PubMed]
- Edgar R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A. J., Van Dongen S., and Ouzounis C. A., 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30: 1575–1584. 10.1093/nar/30.7.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffinet B., Buck W. R., and Shaw A. J., 2009. Morphology, anatomy, and classification of the Bryophyta. Bryo. Biol. 2: 55–138.
- Goodale C. L., Apps M. J., Birdsey R. A., Field C. B., Heath L. S. et al. , 2002. Forest carbon sinks in the northern hemisphere. Ecol. Appl. 12: 891–899. 10.1890/1051-0761(2002)012[0891:FCSITN]2.0.CO;2 [DOI] [Google Scholar]
- Griesmann M., Chang Y., Liu X., Song Y., Haberer G. et al. , 2018. Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science 361: eaat1743. [DOI] [PubMed]
- Haas B. J., Salzberg S. L., Zhu W., Pertea M., Allen J. E. et al. , 2008. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 9: R7 10.1186/gb-2008-9-1-r7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C., and Yandell M., 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12: 491 10.1186/1471-2105-12-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ininbergs K., Bay G., Rasmussen U., Wardle D. A., and Nilsson M.-C., 2011. Composition and diversity of nifH genes of nitrogen-fixing cyanobacteria associated with boreal forest feather mosses. New Phytol. 192: 507–517. 10.1111/j.1469-8137.2011.03809.x [DOI] [PubMed] [Google Scholar]
- Johnson M. G., Malley C., Goffinet B., Shaw A. J., and Wickett N. J., 2016. A phylotranscriptomic analysis of gene family expansion and evolution in the largest order of pleurocarpous mosses (Hypnales, Bryophyta). Mol. Phylogenet. Evol. 98: 29–40. 10.1016/j.ympev.2016.01.008 [DOI] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H.-Y., Fraser M., Li W. et al. , 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30: 1236–1240. 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., and Salzberg S. L., 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D., Ullrich K. K., Murat F., Fuchs J., Jenkins J. et al. , 2018. The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution. Plant J. 93: 515–533. 10.1111/tpj.13801 [DOI] [PubMed] [Google Scholar]
- Li L., Stoeckert C., and Roos D., 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13: 2178–2189. 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindo Z., and Gonzalez A., 2010. The Bryosphere: An integral and influential component of the Earth’s biosphere. Ecosystems (N. Y.) 13: 612–627. 10.1007/s10021-010-9336-3 [DOI] [Google Scholar]
- Lindo Z., Nilsson M.-C., and Gundale M. J., 2013. Bryophyte-cyanobacteria associations as regulators of the northern latitude carbon balance in response to global change. Glob. Change Biol. 19: 2022–2035. 10.1111/gcb.12175 [DOI] [PubMed] [Google Scholar]
- Liu Y., Johnson M. G., Cox C. J., Medina R., Devos N. et al. , 2019. Resolution of the ordinal phylogeny of mosses using targeted exons from organellar and nuclear genomes. Nat. Commun. 10: 1485 10.1038/s41467-019-09454-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M., and Eddy S. R., 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964. 10.1093/nar/25.5.0955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R., Liu B., Xie Y., Li Z., Huang W. et al. , 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1: 18 Erratum: 4: 30. 10.1186/2047-217X-1-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane M., and Consortium U., 2011. UniProt Knowledgebase: a hub of integrated protein data. Database: The Journal of Biological Databases and Curation 2011: bar009. [DOI] [PMC free article] [PubMed]
- Martin J., Bruno V. M., Fang Z., Meng X., Blow M. et al. , 2010. Rnnotator: an automated de novo transcriptome assembly pipeline from stranded RNA-Seq reads. BMC Genomics 11: 663 10.1186/1471-2164-11-663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G., and Kingsford C., 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27: 764–770. 10.1093/bioinformatics/btr011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKain M. R., Wickett N., Zhang Y., Ayyampalayam S., McCombie W. R. et al. , 2012. Phylogenomic analysis of transcriptome data elucidates co-occurrence of a paleopolyploid event and the origin of bimodal karyotypes in Agavoideae (Asparagaceae). Am. J. Bot. 99: 397–406. 10.3732/ajb.1100537 [DOI] [PubMed] [Google Scholar]
- Nawrocki E. P., Burge S. W., Bateman A., Daub J., Eberhardt R. Y. et al. , 2015. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 43: D130–D137. 10.1093/nar/gku1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson M.-C., and Wardle D. A., 2005. Understory vegetation as a forest ecosystem driver: evidence from the northern Swedish boreal forest. Front. Ecol. Environ. 3: 421–428. 10.1890/1540-9295(2005)003[0421:UVAAFE]2.0.CO;2 [DOI] [Google Scholar]
- Pan Y., Birdsey R. A., Phillips O. L., and Jackson R. B., 2013. The structure, distribution, and biomass of the world’s forests. Annu. Rev. Ecol. Evol. Syst. 44: 593–622. 10.1146/annurev-ecolsys-110512-135914 [DOI] [Google Scholar]
- Pertea M., Pertea G. M., Antonescu C. M., Chang T.-C., Mendell J. T. et al. , 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33: 290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing S. A., Lang D., Zimmer A. D., Terry A., Salamov A. et al. , 2008. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69. 10.1126/science.1150646 [DOI] [PubMed] [Google Scholar]
- Schlink K., and Reski R., 2002. Preparing high-quality DNA from moss (Physcomitrella patens). Plant Mol. Biol. Report. 20: 423a–423f. 10.1007/BF02772133 [DOI] [Google Scholar]
- Scrucca L., Fop M., Murphy T. B., and Raftery A. E., 2016. mclust 5: Clustering, Classification and density estimation using Gaussian finite mixture models. R J. 8: 289–317. 10.32614/RJ-2016-021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A. J., Schmutz J., Devos N., Shu S., Carrell A. A. et al. , 2016. Chapter Five - The Sphagnum Genome Project: A new model for ecological and evolutionary genomics, pp. 167–187 in Advances in Botanical Research, edited by Rensing S. A. Academic Press; Cambridge, Massachusetts. [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., and Zdobnov E. M., 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Simpson J. T., Wong K., Jackman S. D., Schein J. E., Jones S. J. et al. , 2009. ABySS: A parallel assembler for short read sequence data. Genome Res. 19: 1117–1123. 10.1101/gr.089532.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A. F., Hubley R., and Green P., 1996-2010 Repeat-Masker Open-3.0. http://www. repeatmasker. Org.
- Stanke M., Diekhans M., Baertsch R., and Haussler D., 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24: 637–644. 10.1093/bioinformatics/btn013 [DOI] [PubMed] [Google Scholar]
- Sun H., Ding J., Piednoël M., and Schneeberger K., 2017. findGSE: estimating genome size variation within human and Arabidopsis using k-mer frequencies. Bioinformatics 34: 550–557. 10.1093/bioinformatics/btx637 [DOI] [PubMed] [Google Scholar]
- Suyama M., Torrents D., and Bork P., 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34: W609–W612. 10.1093/nar/gkl315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turetsky M. R., 2003. The Role of bryophytes in carbon and nitrogen Cycling. Bryologist 106: 395–409. 10.1639/05 [DOI] [Google Scholar]
- Vanneste K., Sterck L., Myburg A. A., Van de Peer Y., and Mizrachi E., 2015. Horsetails are ancient polyploids: evidence from Equisetum giganteum. Plant Cell 27: 1567–1578. 10.1105/tpc.15.00157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H., 2000. Nuclear DNA amounts in Mosses (Musci). Ann. Bot. (Lond.) 85: 531–546. 10.1006/anbo.1999.1103 [DOI] [Google Scholar]
- Warshan D., Bay G., Nahar N., Wardle D. A., Nilsson M.-C. et al. , 2016. Seasonal variation in nifH abundance and expression of cyanobacterial communities associated with boreal feather mosses. ISME J. 10: 2198–2208. 10.1038/ismej.2016.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshan D., Espinoza J. L., Stuart R. K., Richter R. A., Kim S.-Y. et al. , 2017. Feathermoss and epiphytic Nostoc cooperate differently: expanding the spectrum of plant–cyanobacteria symbiosis. ISME J. 11: 2821–2833. 10.1038/ismej.2017.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshan D., Liaimer A., Pederson E., Kim S.-Y., Shapiro N. et al. , 2018. Genomic changes associated with the evolutionary transitions of Nostoc to a plant symbiont. Mol. Biol. Evol. 35: 1160–1175. 10.1093/molbev/msy029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., 2007. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Zerbino D. R., and Birney E., 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18: 821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data are deposited in NCBI with SRA accessions numbers; SRR8297981 and SRR8297982 (BioProject accession number PRJNA509035). The BioSample is available with accession number SAMN10578977 at NCBI. The assembled genome is available with the accession number VACF00000000 at NCBI. All of the annotation files are available at github: https://github.com/PycnopodiaD/Pleurozium_schreberi_annotated_genome_files. Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.7775951.