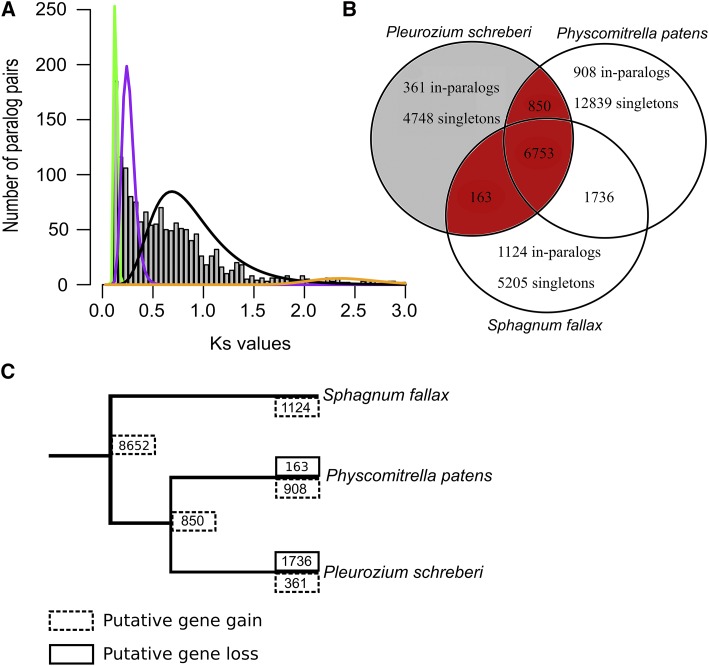
Figure 2.
Detection of whole genome duplication in P. schreberi, shared and unique orthologs, and gene flux between three moss genomes. (A) Distribution of synonymous substitution rates (Ks) among pairs of paralogous genes within P. schreberi. The solid curved lines are the inferred distributions from the mixture model, and colors represent the four component distributions under the model with the highest BIC score. (B) Venn diagram showing shared and unique gene families between moss species. Gene families shared between P. schreberi, P. patens and/or S. fallax are shown in red and the set unique to P. schreberi in gray. (C) Phylogenetic relationships between moss species and inferred gene families gain and loss in this lineage. The numbers of gain/loss events are represented by a solid and dotted box, respectively.