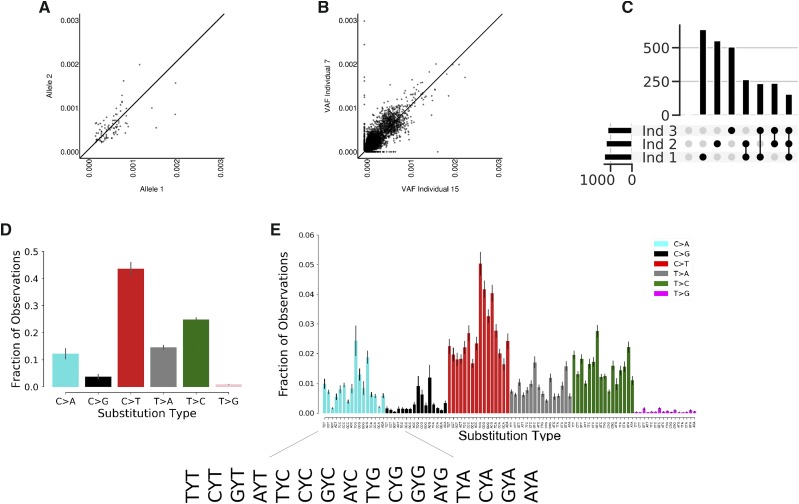
Figure 2.
The background of false positive variants is similar across individuals. A) Using heterozygous SNPs to identify different alleles in human leukocyte gDNA, rare variants within consensus reads equally associate with each of the alleles suggesting they are occurring randomly ex-vivo. Axes are VAFs of variants found on corresponding alleles. B) Using FERMI to measure somatic mutation loads within two different samples shows that background signal is similar within leukocyte gDNA. Axes are VAFs of variants found within each individual. Points represent specific substitutions at each locus. C) The insertions/deletions found within three different individuals were identified. Indel counts are shown on the y-axis. Black dots represent sample groups, where the indel counts are those found within all samples indicated (either each sample alone, or commonly found across the indicated samples; for example, the next to the last vertical bar reflects indels found in both samples 3 and 2). Horizontal bars quantify the total number of indels found in a sample. D) Relative prevalence of observed substitutions within background signal found in leukocyte gDNA consensus reads. Complementary changes such as C > T and G > A are combined. Error is standard deviation across 20 individuals. E) Relative prevalence of observed substitutions classified by the neighboring upstream and downstream nucleotides (trinucleotide context). Error is standard deviation across 20 individuals.