Abstract
Infectious pancreatic necrosis (IPN) is a viral disease with considerable negative impact on the rainbow trout (Oncorhynchus mykiss) aquaculture industry. The aim of the present work was to detect genomic regions that explain resistance to infectious pancreatic necrosis virus (IPNV) in rainbow trout. A total of 2,278 fish from 58 full-sib families were challenged with IPNV and 768 individuals were genotyped (488 resistant and 280 susceptible), using a 57K SNP panel Axiom, Affymetrix. A genome-wide association study (GWAS) was performed using the phenotypes time to death (TD) and binary survival (BS), along with the genotypes of the challenged fish using a Bayesian model (Bayes C). Heritabilities for resistance to IPNV estimated using genomic information, were 0.53 and 0.82 for TD and BS, respectively. The Bayesian GWAS detected a SNP located on chromosome 5 explaining 19% of the genetic variance for TD. The proximity of Sentrin-specific protease 5 (SENP5) to this SNP makes it a candidate gene for resistance against IPNV. In case of BS, a SNP located on chromosome 23 was detected explaining 9% of the genetic variance. However, the moderate-low proportion of variance explained by the detected marker leads to the conclusion that the incorporation of all genomic information, through genomic selection, would be the most appropriate approach to accelerate genetic progress for the improvement of resistance against IPNV in rainbow trout.
Keywords: Bayes C, Oncorhynchus mykiss, Disease resistance, GWAS, QTL, Candidate genes
Infectious pancreatic necrosis (IPN) is a highly transmissible disease caused by IPN virus (IPNV) which belongs to the Birnaviridae family, genus Aquabirnavirus (Roberts and Pearson 2005). This virus affects several wild and cultured aquatic organisms. Salmonid species are especially susceptible to IPNV, thus this disease has a great impact on fish farm operations. The mortality levels during an IPN outbreak are influenced by numerous factors; including species, age of the fish, environmental conditions (Dorson and Touchy 1981), and genetic background, which has been proven to confer resistance to some Atlantic salmon (Salmo salar) (Guy et al. 2006) and rainbow trout (Oncorhynchus mykiss) families (Flores-Mara et al., 2017).
Disease resistance represents a broad term defined as the ability of a host to exert some degree of control over the pathogen life cycle (Bishop and Woolliams 2014). From a quantitative genetics point of view, disease resistance in fish can be measured using survival data from either field tests or controlled experimental challenges (Ødegård et al. 2011, Yáñez et al. 2014). In salmonids it has been possible to determine the presence of significant genetic variation for resistance to bacterial (Gjedrem et al. 1991; Gjedrem and Gjøen 1995; Leeds et al. 2010; Silverstein et al. 2009; Yáñez et al. 2013; Yáñez et al. 2016; Vallejo et al. 2017; Yoshida et al. 2018; Barría et al. 2018), parasitic (Glover et al. 2005; Kolstad et al. 2005; Lhorente et al. 2012; Yáñez et al. 2013; Ødegård et al. 2014), and viral diseases (Guy et al. 2006; Henryon et al. 2005; Ødegård et al. 2007). This implies the possibility of improving, through artificial selection, resistance to various diseases in order to enhance disease control strategies in farmed fish.
Prior to include IPNV resistance as part of the breeding goal it is necessary to determine the genetic variance and heritability for the trait. The current dataset comprises animals that were subjected to an IPNV experimental challenge, as previously reported by Yoshida et al. (2019). However, these authors focused in estimating genetic parameters and evaluating the accuracy of estimated breeding values using pedigree- and genomic-based information. In contrast, the current work provides novel insights of the genetic architecture and the identification of genomic regions associated to the resistance against IPNV in rainbowtrout. The detection of quantitative trait loci (QTL) is the starting point for determining functional variants involved in quantitative traits. In addition, this information could be used to accelerate the improvement of traits through the application of marker assisted selection (MAS) or genomic selection (GS) (Yáñez et al. 2015). The identification of genes that underlie QTL can lead to fundamental knowledge of genetic regulation for disease resistance and host-virus interactions in fish (Moen et al., 2009).
QTL for resistance against various diseases have been determined in salmon and trout (Barría et al., 2018; Kjøglum et al., 2006; Ozaki et al., 2007; Correa et al., 2016; Correa et al., 2015; Moen et al., 2015; Houston et al., 2008; Houston et al., 2012). In Atlantic salmon there is considerable evience for a major QTL for resistance against IPNV in post-smolts, based on data from both field outbreaks (Houston et al., 2008) and experimental challenges (Moen et al., 2009). The discovery of a candidate gene that can bind the virus and successful marker-assited selection for IPNV resistance has also been reported in Atlantic salmon (Moen et al., 2015). However, in rainbow trout there is limited information on the molecular genetic basis of resistance to IPNV. Thus, the aim of the present study was to perform a genome-wide association (GWAS) analysis to determine the genetic architecture for IPNV resistance and to identify potential candidate genes involved in the genetic variation of this trait in rainbow trout.
Materials And Methods
Fish
The population used in this study belonged to the breeding population maintained at Aguas Claras S.A. Experimental tests were conducted at the ATC Patagonia Research Center - Aquainnovo (Puerto Montt, Chile). The fish belonged to 58 full-sib families generated from 58 females and 20 males of rainbow trout from the 2014 year-class. For more details on the description about rearing conditions and population management see Flores-Mara et al. (2017), Neto et al. (2019) and Rodríguez et al. (2018).
Experimental challenge
For a detailed step-by-step protocol for experimental challenge please see Yoshida et al. (2019). Briefly, fish at an average age of 154 (SD = 15) days, weighing an average of 2.24g (SD = 0.71) were kept in a single 0.25 m3 tank under a fresh water recirculation system. The experimental challenge was performed in two steps; i) intraperitoneal injection of inoculum, at a concentration of 107.82 TCID50/mL, using a quantity of 0.05 mL/fish; and ii) immersion pouring 1.1 L of the inoculum plus 5 L of water into the tank containing 130 L of fresh water, maintained at retained flow at 17° for 4 h. Mortality was individually recorded on a daily basis. At day 63 the experiment was stopped and all surviving animals were killed. Fin clip samples were taken from 768 fish and stored in 98% ethanol at -80° until genomic DNA extraction. The phenotypic data for resistance to IPNV were obtained from a total of 2,278 fish.
Genotyping
For the protocol used for genomic DNA extraction, genotyping and quality control (QC) please see Yoshida et al. (2019). Briefly, each fish was genotyped using a 57K Affimetrix Axiom SNP array developed by Palti et al. (2015). The QC included Hardy Weinberg equilibrium, minor allele frequency and call rate for SNPs and samples. The animals and markers which passed the QC were subsequently used for genomic association analysis.
Genome-wide association analysis (GWAS)
The resistance phenotypes were defined as the time to death (TD), measured in days with values ranging from 1 to 63, depending on the day of death; and the binary survival (BS), recorded as 1 if the individual died during the challenge and 0 if the individual survived until the end of the trial. The Bayes C (Habier et al. 2011) analyses were performed using the GS3 software (Legarra et al. 2010). A total of 200,000 iterations were used in the Gibbs sampling, with a burn-in period of 50,000 cycles where results were saved every 50 cycles, totaling 4,000 samples. Convergence and autocorrelation were assessed by visual inspection of trace plots of the posterior variance components. The adjusted model can be described, in matrix notation, as follows:
where y is the vector of phenotypic records for TD or BS; X is an incidence matrix of fixed effects; b is the vector of fixed effect (tagging weight as covariate); Z is an incidence matrix of polygenic effects; u is a random vector of polygenic effects of all individuals in the pedigree; gi is the vector of the genotypes for the ith SNP for each animal; ai is the random allele substitution effect of the ith SNP, is an indicator variable (0, 1) sampled from a binomial distribution with such determined parameters that 1% of the markers were included in the model; and e is a vector of residual effects.
The proportion of the genetic variance explained by each SNP was calculated according to the following formula: where and are the allele frequencies for the i-th SNP; ai is the estimated additive effect of the i-th SNP on the phenotype; and is the estimate of the polygenic variance (Lee et al. 2013).
The association of the SNPs with phenotypes were assessed using Bayes factor (BF), which was calculated as follows: , where p is the posterior probability of a SNP to be assigned a non-zero effect and π = 0.99 is the a priori probability of a SNP to be included in the analysis (Kass & Raftery 1995; Varona et al. 2001).
Candidate genes
The nucleotide sequences surrounding the top ten SNPs that accounted for the largest proportion of genetic variance for each of the IPNV resistance traits were positioned in the most recent version of the rainbow trout reference genome available from NCBI (GenBank assembly Accession GCA_002163495) (Gao et al., 2018; Pearse et al., 2018) using BLASTx (Basic Local Alignment Search Tool) (Altschul et al. 1997). The presence of annotated genes within 1Mb windows surrounding the top ten SNPs was assessed.
Data availability
Phenotype data and genotypes are available at the online repository Figshare.Supplemental material available at FigShare: https://figshare.com/articles/Untitled_Item/7725668.
Results
Experimental challenge and samples
The percentage of accumulated mortality was 13.77%, ranging between 0 to 47.6% for the most and least resistant family, respectively. A selective genotyping strategy was carried out and 768 fish were selected. Table 1 shows the summary statistics of the animals selected for genotyping. For a detailed description about experimental challenge results, please see Yoshida et al. (2019).
Table 1. Summary statistics for the 768 genotyped fish.
| Variable | Mean | Standard deviation | Standard error | Minimum | Maximum |
|---|---|---|---|---|---|
| Weight (g) | 2.12 | 0.74 | 0.03 | 0.80 | 5.80 |
| Time to death (days) | 51.46 | 13.98 | 0.51 | 13.00 | 62.00 |
| Binary survival (0 or 1) | 0.36 | 0.48 | 0.02 | 0.00 | 1.00 |
GWAS and candidate genes
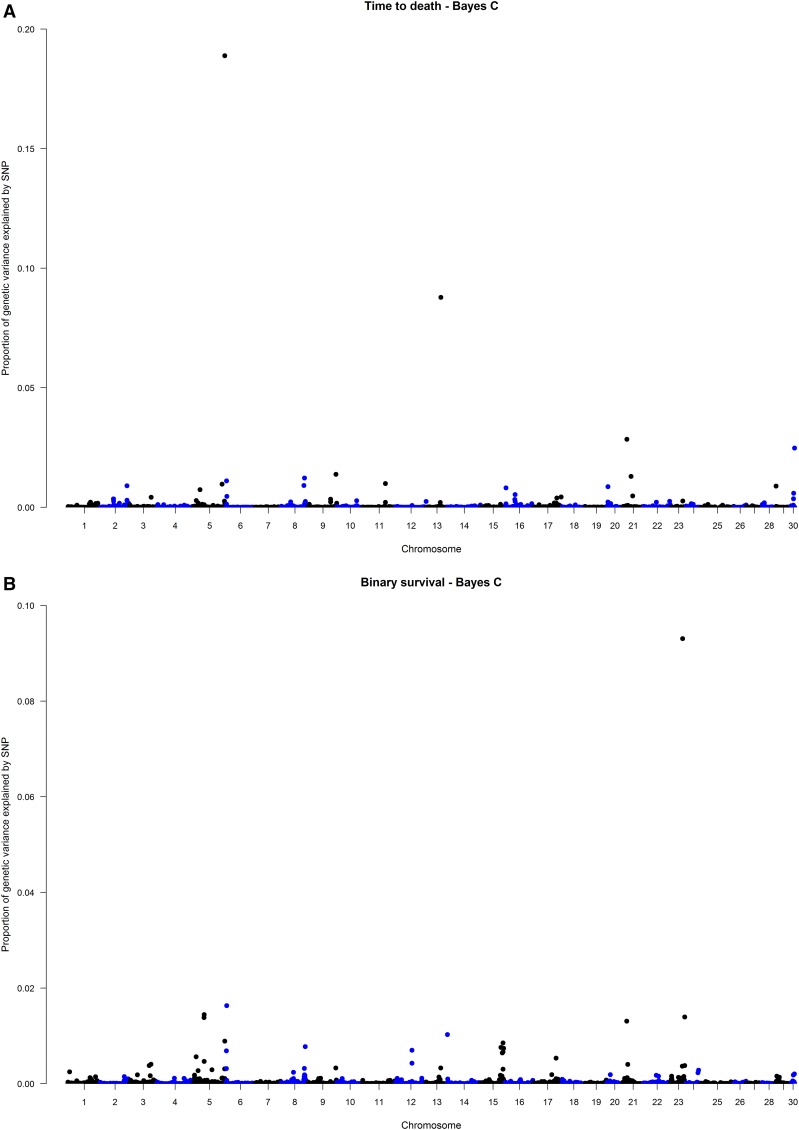
A total of 721 individuals and 38,296 SNP passed QC. The average number of SNPs per chromosome was 1,277 (SD = 384) and varied between 594 and 1,818. The variance components and heritabilities estimated using genomic information are shown in Table 2. Heritabilities were high for both traits (0.53 ± 0.05 and 0.82 ± 0.03 for TD and BS, respectively). The top SNP for both traits explained 18.89% and 9.31% of the genetic variance for TD and BS, respectively. The Figures 1a and 1b show the manhattan plot for both TD and BS, respectively. According to Vidal et al. (2005), a BF from 3 to 20 is suggestive of linkage and BF from 20 to 150 indicates linkage between the SNP and the trait under investigation. In this study, we focused the results and discussion on the SNPs with a BF greater than 20. For TD, the top four SNP that presented a BF greater than 20 are located on chromosomes 5 (AX-89921775), 13 (AX-89964133), 21 (AX-89928391) and 30 (AX-89972475). The sum of these SNPs explained 32.99% of all genetic variance. For the BS trait only the top one SNP presented a BF greater than 20, it is located on chromosome 23 (AX-89938762, Figure 1b) and explained 9.31% of the genetic variance for this trait. The most relevant genes next to the top ten SNPs that explained the highest proportion of genetic variance for each trait are shown in Table 3.
Table 2.
Mean ± standard deviation of variance components estimated using genome-wide SNPs markers for time to death and binary survival for rainbow trout using Bayes C.
| Trait | Total genetic variance1 | Residual variance | Heritability |
|---|---|---|---|
| Time to death | 191.06±139.10 | 78.60±6.78 | 0.53±0.05 |
| Binary Survival | 4.66±1.05 | 1.00±0.00 | 0.82±0.03 |
Correspond to the genetic variance due to markers and the polygenic pedigree-based additive genetic variance.
Figure 1.
Manhattan plot of variance explained by each SNP using Bayes C method. (A) Time to death. (B) Binary survival.
Table 3.
Top 10 SNPs associated with resistance to IPNV in rainbow trout identified using Bayes C model.
| Marker | Chr1 | Position (Mb) | BF2 | Var3 | Gene4,5(6) |
|---|---|---|---|---|---|
| Time to death | |||||
| AX-89921775 | 5 | 86.195 | 93.824 | 0.188 | Sentrin-specific protease 5 (346.42) |
| AX-89964133 | 13 | 18.398 | 43.282 | 0.087 | C-C chemokine receptor type 7 (91.96); |
| Integrin alpha-11-like (239.47); Secretory phospholipase A2 receptor-like protein (279.15) | |||||
| AX-89928391 | 21 | 40.422 | 21.698 | 0.028 | NI7 |
| AX-89972475 | Scaffold | 3.313 | 22.659 | 0.024 | Protein FAM49A ( 228.21) |
| AX-89919030 | 9 | 59.547 | 15.660 | 0.013 | Protein FAM3A (148.87) |
| E3 ubiquitin-protein ligase HUWE1 (373.86) | |||||
| AX-89929860 | 21 | 33.915 | 14.577 | 0.013 | Interleukin-8 (252.85) |
| AX-89919605 | 8 | 15.655 | 14.905 | 0.012 | Laminin subunit alpha-4 (403.02) |
| AX-89964260 | 6 | 71.349 | 13.728 | 0.011 | NI |
| AX-89954884 | 11 | 23.271 | 14.522 | 0.010 | Integrin beta-1 (498.18) |
| AX-89955973 | 5 | 81.679 | 13.401 | 0.009 | Dynamin 1 (83.40) |
| Binary Survival | |||||
| AX-89938762 | 23 | 13.468 | 22.504 | 0.093 | Apoptosis-stimulating of p53 protein 2-like ( 274.14) |
| AX-89960378 | 6 | 69.996 | 9.787 | 0.016 | NI |
| AX-89950201 | 5 | 29.774 | 9.274 | 0.014 | Myosin-IIIb-like (60.73) |
| Protein FAM78A-like (418.01) | |||||
| AX-89951900 | 23 | 8.146 | 9.208 | 0.014 | NI |
| AX-89944302 | 5 | 29.773 | 8.910 | 0.013 | Myosin-IIIb-like (60.41) |
| Protein FAM78A-like (417.69) | |||||
| AX-89928391 | 21 | 40.422 | 8.618 | 0.013 | NI |
| AX-89952306 | 14 | 0.420 | 8.264 | 0.010 | NI |
| AX-89946704 | 5 | 86.995 | 7.395 | 0.008 | Lamin-B2 ( 351.97) |
| AX-89933583 | 15 | 13.021 | 7.392 | 0.008 | E3 ubiquitin-protein ligase RNF31-like (377.72); Interferon regulatory factor 4-like (337.82); Signalosome complex subunit 5 (428.51) |
| AX-89962297 | 8 | 12.919 | 7.680 | 0.007 | Unconventional myosin-VI-like (38.04); Collagen alpha-1(XII) chain-like (195.72); Sentrin-specific protease 6 (73.33) |
Chromosome.
Bayes factor.
Proportion of variance explained by each individual marker.
Suggestive candidate genes located within 1-Mb window.
Oncorhynchus mykiss used as reference species.
Proximity to the SNP (kbp)
Not identified
Discussion
Experimental challenge and heritability
IPN is one of the main diseases affecting salmonid aquaculture. Our current work was done using data obtained from an experimental challenge to IPNV previously presented by Yoshida et al. (2019). Nonetheless, previous authors focused on determining the improvement of genomic prediction accuracies using a single step approach (including genotyped and non-genotyped animals) for the genetic evaluations of resistance to IPNV. The current study focused on providing novel insights of the genetic architecture of IPNV resistance and the genomic regions involved in this trait in a farmed rainbow trout population. The animals used in this study were genotyped using a selective genotyping approach. The allocation of genotyping resources was directed to the most genetically informative progeny (extremely high and low phenotypic values). The selective genotype is a common strategy used to reduce genotyping costs and it has been shown to be highly suitable for QTL detection in genome-wide association studies in livestock and aquaculture species.
The mortality rate in IPNV challenge experiments is highly variable and depends on several factors, including the virus strain, the population evaluated, the size of the fish, environmental conditions, among others. For example, mortality rates over 40% have been reported for Atlantic salmon (Roberts and Pearson 2005; Houston et al. 2008). Other authors have reported mortality rates of 70.5% and 77.8% for the same disease and species (Moen et al. 2009), while in rainbow trout mortality rates range from 36 to 54% (Ozaki et al.,2007).
Some studies have shown significant pedigree-based heritabilities for IPNV resistance in rainbow trout and Atlantic salmon (Flores-Mara et al. 2017; Guy et al. 2006; Storset et al. 2007) using survival time as the trait definition. For instance, in Atlantic salmon, heritability values ranging between 0.26 and 0.55 have been reported for resistance to IPNV, measured as binary survival and analyzed by threshold models (Wetten et al. 2007; Kjøglum et al. 2008; Guy et al. 2009; Gheyas et al. 2010; Houston et al. 2010). Previous studies have reported pedigree-based heritability values of 0.39 and 0.4 for time to death and 0.35 for binary survival in the same population of IPNV-infected rainbow trout (Flores-Mara et al. 2017; Yoshida et al. 2019). Moreover, heritability values of 0.24 and 0.25 for day to death and binary survival, respectively, have been reported (Yoshida et al. 2019) using a combination of both the pedigree relationship matrix (A) and the genomic matrix (G), also called the H matrix (Aguilar et al. 2010). The heritability values calculated in the present study using genomic information are higher than values previously reported in rainbow trout and indicate that selection to improve resistance to IPNV is feasible. The proportion of genetic variance explained by the SNP with the largest effect was larger for trait TD than the SNP identified for trait BS.
GWAS and candidate genes
Previous studies in Atlantic salmon have determined two significant QTL for resistance against IPNV. The most significant one, explaining 29% and 83% of the phenotypic and genetic variance, respectively, was identified on chromosome 26 (Houston et al. 2008; 2012). This QTL was confirmed in an independent Atlantic salmon population from Norway (Moen et al. 2009). In rainbow trout, Ozaki et al. (2001) found 2 QTL associated with resistance to IPNV using the linkage map elaborated by Sakamoto et al. (2000) and based on microsatellite markers. The first QTL was found in linkage group 3 while the second QTL was identified in linkage group 22, each of which explains about 17% of the phenotypic variance (Ozaki et al. 2001). The same QTL were confirmed in a subsequent study (Ozaki et al. 2007), in which another significant QTL was detected in linkage group 12. Based on the linkage map developed by Palti et al. (2012) these significant markers were located on chromosome 14 and 16, respectively (Hu et al. 2016).
Some factors could influence the QTL detection, such as the genetic architecture of the traits, the number of animals genotyped and phenotyped, trait’s heritability, linkage disequilibrium (LD) between markers and QTL, and models for QTL detection (Van den Berg et al. 2013). The LD is dependent on the demographic history, effective population size, among others, whereas heritability is a property of the trait, population and environmental condition in which the trait was measured (Falconer and Mackay 1996), suggesting that these parameters are intrinsic for a specific population. In our study we did not identify QTL with major effects, similar to those found by Houston et al. (2012) and Moen et al. (2015) in Atlantic salmon. Moreover, none of the QTL presented here were nearby the known location of the epithelial cadherin gene in the rainbow trout genome. A recent study compared the genomic regions associated to the resistance against a bacterial infection in three salmonid species (O. kisutch, S. salar and O. mykiss), indicating that there is a reduced overlapping between the particular genes involved in the trait among species (Yáñez et al. 2019).
The SNP that explained the greatest proportion of genetic variance for TD in the present study (AX-89921775) is located on chromosome 5 (Figure 1a), in a region that contains a gene that encodes Sentrin-specific protease 5. In mammals, this protein participates in the SUMO pathway (small ubiquitin-like modifier). Its function is mainly related to the activity of isopeptidase (Pinto et al. 2012) and the regulation of biological processes that are key to viral replication, including genetic transcription, cell cycle, apoptosis, intracellular and intranuclear trafficking, and protein stability.
Resistance to infection can be the result of a more robust host immune response to the invading agent. In this study we identified SNP located near several genes that encode proteins whose function is to control inflammation as well as other immune-related responses, including the gene that encodes Secretory phospholipase A2 receptor-like protein (close to SNP AX-89964133 in chromosome 13). Phospholipases A2 are enzymes released in plasma and extracellular fluids during inflammatory diseases, that induces the release of the pro-inflammatory cytokines TNF-α and IL-6 depending on the concentration of protein (Granata et al. 2005).
Thr SNP AX-89933583 (chromosome 15) is associated to BS, which is located in a region that encodes interferon regulatory factor 4-like (irf4). This protein is important in both the regulation of interferons in response to virus infection and the regulation of inducible genes by interferon. In fact, in mammals irf4 plays a central role in TH cell regulation (Mahnke et al. 2016), a fundamental cell-mediated process for the host antiviral response. The SNP AX-89933583 is also located in a gene region that encodes for signalosome complex subunit 5-like, that is involved in the regulation of nuclear factor kappa B (NF-kB) one of the most relevant transcription factors involved in the control of several pro-inflammatory proteins (Adcock and Caramori 2001).
The activation of the immune response also requires the expression of genes responsible for the activation and recruitment of immune cell populations to the site of infection. In our study, SNP AX-89929860 was located in a gene region that codes for Interleukin-8 (IL-8), that is a member of the CXC chemokine subfamily directly associated with the pro-inflammatory response because it attracts different immune cell populations including neutrophils, T lymphocytes and basophils (Reyes-Cerpa et al. 2012). Importantly, an increased gene expression for Interleukin-8 was observed in symptomatic IPNV-infected head kidney trout but not in fish with persistent asymptomatic infection (Reyes-Cerpa et al. 2014), suggesting its potential effect on the inflammatory response. SNP AX-89954884 is located near integrin beta 1; which is a membrane receptor involved in cell adhesion and recognition in a variety of processes including the immune response, indicating a potential role on influencing the pro-inflammatory response on IPNV-infected fish.
Previous reports in Atlantic salmon have shown clear differences in the gene expression pattern between IPNV-susceptible and –resistant phenotypes showing differences in ubiquitin-dependent and apoptosis processes (Reyes-López et al. 2015; Robledo et al. 2016). In our study two SNPs (AX-89919030; AX-89938762) were located in regions that contain genes that encode for E3 ubiquitin-protein ligase HUWE1 and Apoptosis-stimulating of p53 protein 2-like, respectively. E3 ubiquitin-protein ligase HUWE1 mediates the proteasomal degradation of target proteins, and the results presented here provide more evidence that ubiquitin-dependent processes could be important factors in IPNV disease resistance. The role of HUWE1 in proteasomal degradation of target proteins opens the possibility that other processes associated with this function, like vesicular trafficking, could also be involved with susceptibility to IPNV.
The results obtained from this study indicate that the QTL involved in IPNV resistance contribute to a moderate-low proportion of the variance of this trait. From a practical prespective, the implementation of this information in MAS is probably not the most efficient approach (Correa et al. 2015).
Conclusions
This is the first work reporting the detection and position of QTL involved in IPNV resistance in rainbow trout using SNP markers. Resistance to IPNV can be described as a moderately oligogenic trait since there are probably several loci involved; each with a moderate-low effect. Potential candidate genes have been identified close to the associated SNPs, whose biological role in the fish immune response suggest they could be involved in the mechanisms of resistance against IPNV.
Acknowledgments
The authors would like to thank the company Aguas Claras S.A. for providing the fish and data used in the present study. This study has been partially funded by a CORFO grant (12PIE-17669), Government of Chile. Francisco H. Rodríguez and Raúl Flores-Mara would like to thank the grant Presidente de la República of the Government of Peru.
Footnotes
Supplemental material available at FigShare: https://figshare.com/articles/Untitled_Item/7725668.
Communicating editor: R. Houston
Literature Cited
- Adcock I. M., and Caramori G., 2001. Cross-talk between pro-inflammatory transcription factors and glucocorticoids. Immunol. Cell Biol. 79: 376–384. 10.1046/j.1440-1711.2001.01025.x [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z.. et al, 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar I., Misztal I., Johnson D. L., Legarra A., Tsuruta S.. et al., 2010. Hot topic: A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J. Dairy Sci. 93: 743–752. http://www.sciencedirect.com/science/article/pii/S0022030210715174. 10.3168/jds.2009-2730 [DOI] [PubMed] [Google Scholar]
- Barría A., Christensen K. A., Yoshida G. M., Correa K., Jedlicki A. M.. et al., 2018. Genomic Predictions and Genome-Wide Association Study of Resistance Against Piscirickettsia salmonis in Coho Salmon (Oncorhynchus kisutch) Using ddRAD Sequencing. G3: (Bethesda) 8: 1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop S. C., and Woolliams J. A., 2014. Genomics and disease resistance studies in livestock. Livest. Sci. 166: 190–198. 10.1016/j.livsci.2014.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers R. M., Lapatra S. E., and Dhar A. K., 2008. Detection and quantitation of infectious pancreatic necrosis virus by real-time reverse transcriptase-polymerase chain reaction using lethal and non-lethal tissue sampling. J. Virol. Methods 147: 226–234. 10.1016/j.jviromet.2007.09.003 [DOI] [PubMed] [Google Scholar]
- Correa K., Maass A., Lhorente J. P., López M. E., Bassini L.. et al., 2015. Genome-wide association analysis reveals loci associated with resistance against Piscirickettsia salmonis in two Atlantic salmon (Salmo salar L.) chromosomes. BMC Genomics 16: 854 10.1186/s12864-015-2038-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa, K., J. P. Lhorente, L. Bassini, M. E. López, A. DI Genova et al, 2016 Genome wide association study for resistance to Caligus rogercresseyi in Atlantic salmon (Salmo salar L.) using a 50K SNP genotyping array. Aquaculture.
- Danzmann R. G., Cairney M., Davidson W. S., Ferguson M. M., Gharbi K.. et al., 2005. A comparative analysis of the rainbow trout genome with 2 other species of fish (Arctic charr and Atlantic salmon) within the tetraploid derivative Salmonidae family (subfamily: Salmoninae). Genome 48: 1037–1051. 10.1139/g05-067 [DOI] [PubMed] [Google Scholar]
- Dorson M., and Touchy C., 1981. The influence of fish age and water temperature on mortalities of rainbow trout, Salmo gairdneri Richardson, caused by a European strain of infectious pancreatic necrosis virus. J. Fish Dis. 4: 213–221. 10.1111/j.1365-2761.1981.tb01128.x [DOI] [Google Scholar]
- Falconer D. S., and Mackay T. F. C., 1996. Introduction to Quantitative Genetics, Harlow, UK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Mara R., Rodríguez F. H., Bangera R., Lhorente J. P., Neira R.. et al., 2017. Resistance against infectious pancreatic necrosis exhibits significant genetic variation and is not genetically correlated with harvest weight in rainbow trout (Oncorhynchus Mykiss). Aquaculture 479: 155–160. 10.1016/j.aquaculture.2017.05.042 [DOI] [Google Scholar]
- Gao G., Nome T., Pearse D. E., Moen T., Naish K. A.. et al., 2018. A new single nucleotide polymorphism database for rainbow trout generated through whole genome resequencing. Front. Genet. 9: 147 10.3389/fgene.2018.00147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genabel Project Developers, 2015 GenABEL Tutorial. [Accessed May 2, 2017]. 10.5281/zenodo.19738 [DOI]
- Gheyas A. A., Haley C. S., Guy D. R., Hamilton A., Tinch A. E.. et al., 2010. Effect of a major QTL affecting IPN resistance on production traits in Atlantic salmon. Anim. Genet. 41: 666–668. 10.1111/j.1365-2052.2010.02051.x [DOI] [PubMed] [Google Scholar]
- Gjedrem T., Salte R., and Gjøen H. M., 1991. Genetic variation in susceptibility of Atlantic salmon to furunculosis. Aquaculture 97: 1–6. 10.1016/0044-8486(91)90274-B [DOI] [Google Scholar]
- Gjedrem T., and Gjøen H., 1995. Genetic variation in susceptibility of Atlantic salmon, Salmo salar L., to furunculosis, BKD and cold water vibriosis. Aquacult. Res. 26: 129–134. 10.1111/j.1365-2109.1995.tb00892.x [DOI] [Google Scholar]
- Glover K., Aasmundstad T., Nilsen F., Storset A., and Skaala Ø., 2005. Variation of Atlantic salmon families (Salmo salar L.) in susceptibility to the sea lice Lepeophtheirus salmonis and Caligus elongatus. Aquaculture 245: 19–30. 10.1016/j.aquaculture.2004.11.047 [DOI] [Google Scholar]
- Goddard M. E., and Hayes B. J., 2007. Genomic selection. J. Anim. Breed. Genet. 124: 323–330. 10.1111/j.1439-0388.2007.00702.x [DOI] [PubMed] [Google Scholar]
- Granata F., Petraroli A., Boilard E., Bezzine S., Bollinger J.. et al., 2005. Activation of cytokine production by secreted phospholipase A2 in human lung macrophages expressing the M-type receptor. J. Immunol. 174: 464–474. 10.4049/jimmunol.174.1.464 [DOI] [PubMed] [Google Scholar]
- Guy D. R., Bishop S. C., Brotherstone S., Hamilton A., Roberts R. J.. et al., 2006. Analysis of the incidence of infectious pancreatic necrosis mortality in pedigreed Atlantic salmon, Salmo salar L., populations. J. Fish Dis. 29: 637–647. 10.1111/j.1365-2761.2006.00758.x [DOI] [PubMed] [Google Scholar]
- Guy D. R., Bishop S. C., Woolliams J. A., and Brotherstone S., 2009. Genetic parameters for resistance to infectious pancreatic necrosis in pedigreed Atlantic salmon (Salmo salar) post-smolts using a Reduced Animal Model. Aquaculture 290: 229–235. 10.1016/j.aquaculture.2009.02.015 [DOI] [Google Scholar]
- Habier D., Fernando R. L., Kizilkaya K., and Garrick D. J., 2011. Extension of the bayesian alphabet for genomic selection. BMC Bioinformatics 12: 186 http://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471–2105–12–186. 10.1186/1471-2105-12-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henryon M., Berg P., Olesen N. J., Kjær T. E., Slierendrecht W. J.. et al., 2005. Selective breeding provides an approach to increase resistance of rainbow trout (Oncorhynchus mykiss) to the diseases, enteric redmouth disease, rainbow trout fry syndrome, and viral haemorrhagic septicaemia. Aquaculture 250: 621–636. 10.1016/j.aquaculture.2004.12.022 [DOI] [Google Scholar]
- Houston R. D., Haley C. S., Hamilton A., Guy D. R., Tinch A. E.. et al., 2008. Major Quantitative Trait Loci Affect Resistance to Infectious Pancreatic Necrosis in Atlantic Salmon (Salmo salar). Genetics 178: 1109–1115. 10.1534/genetics.107.082974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston R. D., C. S. Haley, A. Hamilton, D. R. Guy, J. C. Mota-Velasco et al., 2010 The susceptibility of Atlantic salmon fry to freshwater infectious pancreatic necrosis is largely explained by a major QTL. Heredity 105: 318–327. [DOI] [PubMed] [Google Scholar]
- Houston R. D., Davey J. W., Bishop S. C., Lowe N. R., Mota-Velasco J. C.. et al., 2012. Characterisation of QTL-linked and genome-wide restriction site-associated DNA (RAD) markers in farmed Atlantic salmon. BMC Genomics 13: 244 10.1186/1471-2164-13-244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z. L., Park C. A., and Reecy J. M., 2016. Developmental progress and current status of the Animal QTLdb. Nucleic Acids Res. 44: D827–D833. 10.1093/nar/gkv1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass R. E., and Raftery A. E., 1995. Bayes Factors. J. Am. Stat. Assoc. 90: 773–795 http://www.tandfonline.com/doi/abs/10.1080/01621459.1995.10476572. 10.1080/01621459.1995.10476572 [DOI] [Google Scholar]
- Kjøglum S., Larsen S., Bakke H. G., and Grimholt U., 2006. How specific MHC class I and class II combinations affect disease resistance against infectious salmon anaemia in Atlantic salmon (Salmo salar). Fish Shellfish Immunol 21: 431–441. [DOI] [PubMed] [Google Scholar]
- Kjøglum S., Henryon M., Aasmundstad T., and Korsgaard I., 2008. Selective breeding can increase resistance of Atlantic salmon to furunculosis, infectious salmon anaemia and infectious pancreatic necrosis. Aquacult. Res. 39: 498–505. 10.1111/j.1365-2109.2008.01904.x [DOI] [Google Scholar]
- Kolstad K., Heuch P. A., Gjerde B., Gjedrem T., and Salte R., 2005. Genetic variation in resistance of Atlantic salmon (Salmo salar) to the salmon louse Lepeophtheirus salmonis. Aquaculture 247: 145–151. 10.1016/j.aquaculture.2005.02.009 [DOI] [Google Scholar]
- Lee S. H., Choi B. H., Lim D., Gondro C., Cho Y. M.. et al., 2013. Genome-Wide Association Study Identifies Major Loci for Carcass Weight on BTA14 in Hanwoo (Korean Cattle). PLoS One 8: e74677 10.1371/journal.pone.0074677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds T. D., Silverstein J. T., Weber G. M., Vallejo R. L., Palti Y.. et al., 2010. Response to selection for bacterial cold water disease resistance in rainbow trout. J. Anim. Sci. 88: 1936–1946. 10.2527/jas.2009-2538 [DOI] [PubMed] [Google Scholar]
- Legarra, A., A. Ricard, F. Olivier, and I. Flutre, 2010 GS3–Genomic selection, Gibbs sampling, Gauss Seidel and BayesCπ. Available at: https://github.com/alegarra/gs3 [Accessed December 12, 2016].
- Lhorente J. P., Gallardo J. A., Villanueva B., Araya A. M., Torrealba D. A.. et al., 2012. Quantitative genetic basis for resistance to Caligus rogercresseyi sea lice in a breeding population of Atlantic salmon (Salmo salar). Aquaculture 324–325: 55–59. 10.1016/j.aquaculture.2011.10.046 [DOI] [Google Scholar]
- Neto R. V. R., Yoshida G. M., Lhorente J. P., and Yáñez J. M., 2019. Genome-wide association analysis for body weight identifies candidate genes related to development and metabolism in rainbow trout (Oncorhynchus mykiss). Mol. Genet. Genomics 294: 563–571. 10.1007/s00438-018-1518-2 [DOI] [PubMed] [Google Scholar]
- Mahnke J., Schumacher V., Ahrens S., Käding N., Feldhoff L. M.. et al., 2016. Interferon Regulatory Factor 4 controls TH1 cell effector function and metabolism. Sci. Rep. 6: 35521 10.1038/srep35521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen T., Baranski M., Sonesson A. K., and Kjøglum S., 2009. Confirmation and fine-mapping of a major QTL for resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar): population-level associations between markers and trait. BMC Genomics 10: 368 10.1186/1471-2164-10-368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen T., Torgersen J., Santi N., Davidson W. S., Baranski M.. et al., 2015. Epithelial cadherin determines resistance to infectious pancreatic pecrosis virus in atlantic salmon. Genetics 200: 1313–1326. 10.1534/genetics.115.175406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki A., Sakamoto T., Khoo S., Nakamura K., Coimbra M. R.. et al., 2001. Quantitative trait loci (QTLs) associated with resistance/susceptibility to infectious pancreatic necrosis virus (IPNV) in rainbow trout (Oncorhynchus mykiss). Mol. Genet. Genomics 265: 23–31. 10.1007/s004380000392 [DOI] [PubMed] [Google Scholar]
- Ozaki A., Khoo S. K., Yoshiura Y., Ototake M., Sakamoto T.. et al., 2007. Identification of Additional Quantitative Trait Loci (QTL) Responsible for Susceptibility to Infectious Pancreatic Necrosis Virus in Rainbow Trout. Mol. Genet. Genomics 265: 23–31. [DOI] [PubMed] [Google Scholar]
- Ødegård J., Olesen I., Gjerde B., and Klemetsdal G., 2007. Positive genetic correlation between resistance to bacterial (furunculosis) and viral (infectious salmon anaemia) diseases in farmed Atlantic salmon (Salmo salar). Aquaculture 271: 173–177. 10.1016/j.aquaculture.2007.06.006 [DOI] [Google Scholar]
- Ødegård J., Baranski M., Gjerde B., and Gjedrem T., 2011. Methodology for genetic evaluation of disease resistance in aquaculture species: Challenges and future prospects. Aquacult. Res. 42: 103–114. 10.1111/j.1365-2109.2010.02669.x [DOI] [Google Scholar]
- Ødegård J., Moen T., Santi N., Korsvoll S. A., Kjøglum S.. et al., 2014. Genomic Prediction in an Admixed Population of Atlantic Salmon (Salmo Salar). Front. Genet. 5: 1–8. 10.3389/fgene.2014.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palti Y., Genet C., Gao G., Hu Y., You F. M.. et al., 2012. A Second Generation Integrated Map of the Rainbow Trout (Oncorhynchus mykiss) Genome: Analysis of Conserved Synteny with Model Fish Genomes. Mar. Biotechnol. (NY) 14: 343–357. 10.1007/s10126-011-9418-z [DOI] [PubMed] [Google Scholar]
- Palti Y., Gao G., Liu S., Kent M. P., Lien S.. et al., 2015. The development and characterization of a 57K single nucleotide polymorphism array for rainbow trout. Mol. Ecol. Resour. 15: 662–672. 10.1111/1755-0998.12337 [DOI] [PubMed] [Google Scholar]
- Pearse D. E., Barson N. J., Nome T., Gao G., Campbell M. A.. et al., 2018. Sex-dependent dominance maintains migration supergene in rainbow trout. bioRxiv 10.1101/504621. [DOI] [PubMed] [Google Scholar]
- Pinto M. P., Carvalho A. F., Grou C. P., Rodríguez-borges J. E., Sá-Miranda C.. et al., 2012. Heat shock induces a massive but differential inactivation of SUMO-specific proteases. Biochim. Biophys. Acta 1823: 1958–1966. 10.1016/j.bbamcr.2012.07.010 [DOI] [PubMed] [Google Scholar]
- Phillips R. B., Keatley K. A., Morasch M. R., Ventura A. B., Lubieniecki K. P.. et al., 2009. Assignment of Atlantic salmon (Salmo salar). linkage groups to specific chromosomes : Conservation of large syntenic blocks corresponding to whole chromosome arms in rainbow trout (Oncorhynchus mykiss). BMC Genet. 11: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Cerpa S., Maisey K., Reyes-López F., Toro-Ascuy D., Sandino A. M.. et al., 2012. Fish cytokines and immune response, in New Advances and Contributions to Fish Biology. Rijeka, Croatia: Intech. p 3–57. 10.5772/53504 [DOI] [Google Scholar]
- Reyes-Cerpa S., Reyes-López F., Toro-Ascuy D., Montero R., Maisey K.. et al., 2014. Induction of anti-inflammatory cytokine expression by IPNV in persistent infection. Fish Shellfish Immunol. 41: 172–182. 10.1016/j.fsi.2014.08.029 [DOI] [PubMed] [Google Scholar]
- Reyes-López, F. E., E. Vallejos-Vidal, S. Reyes-Cerpa, A. M. Sandino, L. Tort et al., 2015 Differential immune gene expression profiles in susceptible and resistant full-sibling families of Atlantic salmon (Salmo salar) challenged with infectious pancreatic necrosis virus (IPNV). Developmental and Comparative Immunology, 53(1), 210–221. 10.1016/j.dci.2015.06.017 [DOI] [PubMed]
- Roberts R. J., and Pearson M. D., 2005. Infectious pancreatic necrosis in Atlantic salmon, Salmo salar L. J. Fish Dis. 28: 383–390. 10.1111/j.1365-2761.2005.00642.x [DOI] [PubMed] [Google Scholar]
- Robledo D., Taggart J. B., Ireland J. H., McAndrew B. J., Starkey W. G.. et al., 2016. Gene Expression Comparison of Resistant and Susceptible Atlantic Salmon Fry Challenged with Infectious Pancreatic Necrosis Virus Reveals a Marked Contrast in Immune Response. BMC Genomics 17: 279 10.1186/s12864-016-2600-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez F. H., Cáceres G., Lhorente J. P., Newman S., Bangera R.. et al., 2018. Genetic (co)variation in skin pigmentation patterns and growth in rainbow trout. Animal 13: 675–682. 10.1017/S175173111800188X [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Danzmann R. G., Gharbi K., Howard P., Ozaki A.. et al., 2000. A microsatellite linkage map of rainbow trout (oncorhynchus mykiss) characterized by large sex – specific differences in recombination rates. Genetics 155: 1331–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein J. T., Vallejo R. L., Palti Y., Leeds T. D., Rexroad C. E.. et al., 2009. Rainbow trout resistance to bacterial cold-water disease is moderately heritable and is not adversely correlated with growth. J. Anim. Sci. 87: 860–867. 10.2527/jas.2008-1157 [DOI] [PubMed] [Google Scholar]
- Storset A., Strand C., Wetten M., Kjøglum S., and Ramstad A., 2007. Response to selection for resistance against infectious pancreatic necrosis in Atlantic salmon (Salmo salar L.). Aquaculture 272: S62–S68. 10.1016/j.aquaculture.2007.08.011 [DOI] [Google Scholar]
- Varona L., García-Cortés L., and Pérez-Enciso M., 2001. Bayes factors for detection of Quantitative Trait Loci. Genet. Sel. Evol. 33: 133 http://www.gsejournal.org/content/33/2/133. 10.1186/1297-9686-33-2-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo R. L., Liu S., Gao G., Fragomeni B. O., Hernandez A. G.. et al., 2017. Similar Genetic Architecture with Shared and Unique Quantitative Trait Loci for Bacterial Cold Water Disease Resistance in Two Rainbow Trout Breeding Populations. Front. Genet. 8: 156 10.3389/fgene.2017.00156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal, O., J. L. Noguera, M. Amills, L. Varona, M. Gil et al, 2005 Identification of carcass and meat quality quantitative trait loci in a Landrace pig population selected for growth and leanness. Journal of Animal Science, 83:293. Available at: https://www.animalsciencepublications.org/publications/jas/abstracts/83/2/0830293 [Accessed December 6, 2016]. [DOI] [PubMed]
- Wetten M., Aasmundstad T., Kjøglum S., and Storset A., 2007. Genetic analysis of resistance to infectious pancreatic necrosis in Atlantic salmon (Salmo salar L.). Aquaculture 272: 111–117. 10.1016/j.aquaculture.2007.08.046 [DOI] [Google Scholar]
- Yáñez J. M., Bangera R., Lhorente J. P., Oyarzún M., and Neira R., 2013. Quantitative genetic variation of resistance against Piscirickettsia salmonis in Atlantic salmon (Salmo salar). Aquaculture 414–415: 155–159. 10.1016/j.aquaculture.2013.08.009 [DOI] [Google Scholar]
- Yáñez, J. M., S. Naswa, M. E. López, L. Bassini, M. E. Cabrejos et al., 2014 Development of a 200K SNP Array for Atlantic Salmon : Exploiting across continents genetic variation. 10th world congress of genetics applied to livestock production development, (February 2016). [Google Scholar]
- Yáñez J. M., Newman S., and Houston R. D., 2015. Genomics in Aquaculture to Better Understand Species Biology and Accelerate Genetic Progress. Front. Genet. 6: 128 10.3389/fgene.2015.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez, J. M., R. Bangera, J. P. Lhorente, A. Barría, M. Oyarzún et al., 2016 Negative Genetic Correlation between Resistance against Piscirickettsia Salmonis and Harvest Weight in Coho Salmon (Oncorhynchus Kisutch). Aquaculture 459. Elsevier B.V.: 8–13. 10.1016/j.aquaculture.2016.03.020 [DOI]
- Yáñez, J. M, G. M. Yoshida, A. Parra, K. Correa, A. Barria A. et al, 2019 Comparative genomic analysis of three salmonid species identifies functional candidate genes involved in resistance to the intracellular bacterium Piscirickettsia salmonis. 10.3389/fgene.2019.00665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida G. M., Bangera R., Carvalheiro R., Correa K., Figueroa R.. et al., 2018. Genomic Prediction Accuracy for Resistance Against Piscirickettsia Salmonis in Farmed Rainbow Trout. Genes Genomes Genetics 8: 719–726. 10.1534/g3.117.300499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida G. M., Carvalheiro R., Rodríguez F. H., Lhorente J. P., and Yáñez J. M., 2019. Single-step genomic evaluation improves accuracy of breeding value predictions for resistance to infectious pancreatic necrosis virus in rainbow trout. Genomics 111: 127–132. 10.1016/j.ygeno.2018.01.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Phenotype data and genotypes are available at the online repository Figshare.Supplemental material available at FigShare: https://figshare.com/articles/Untitled_Item/7725668.