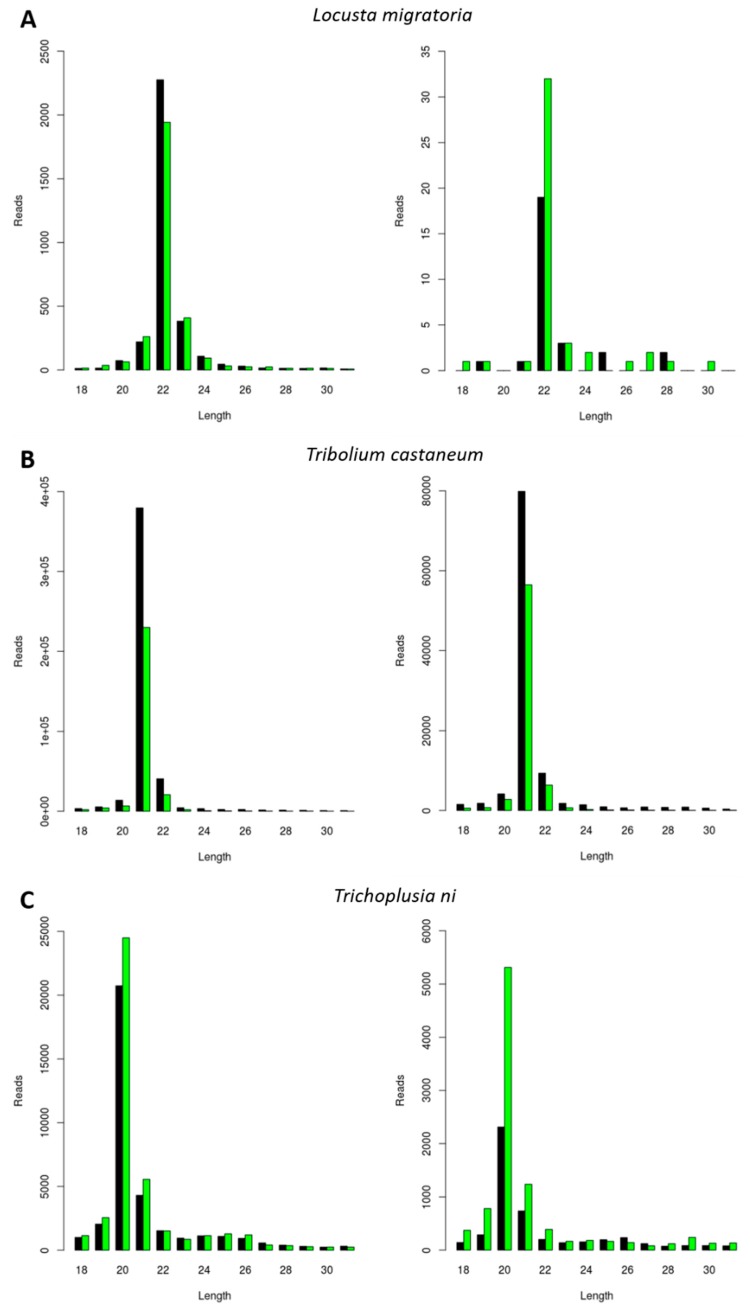
Figure 2.
Generation of Flock House Virus (FHV)-derived siRNAs with species-dependent length distribution. Length distribution of small RNAs mapping to FHV, derived from fat body of FHV-infected L. migratoria (A), as well as from FHV-infected Tribolium castaneum TcA cells (B) and from persistently FHV-infected Trichoplusia ni High Five cells (C). Left: FHV RNA1. Right: FHV RNA2. Reads (y axis): number of reads. Length (x axis): length (nt) of the reads. Black: sense reads. Green: antisense reads. To avoid degradation products and general contaminants, reads shorter than 18 nt and longer than 31 nt were not included. Mapping was performed with Bowtie2, and graphs were obtained with the viRome package in R.