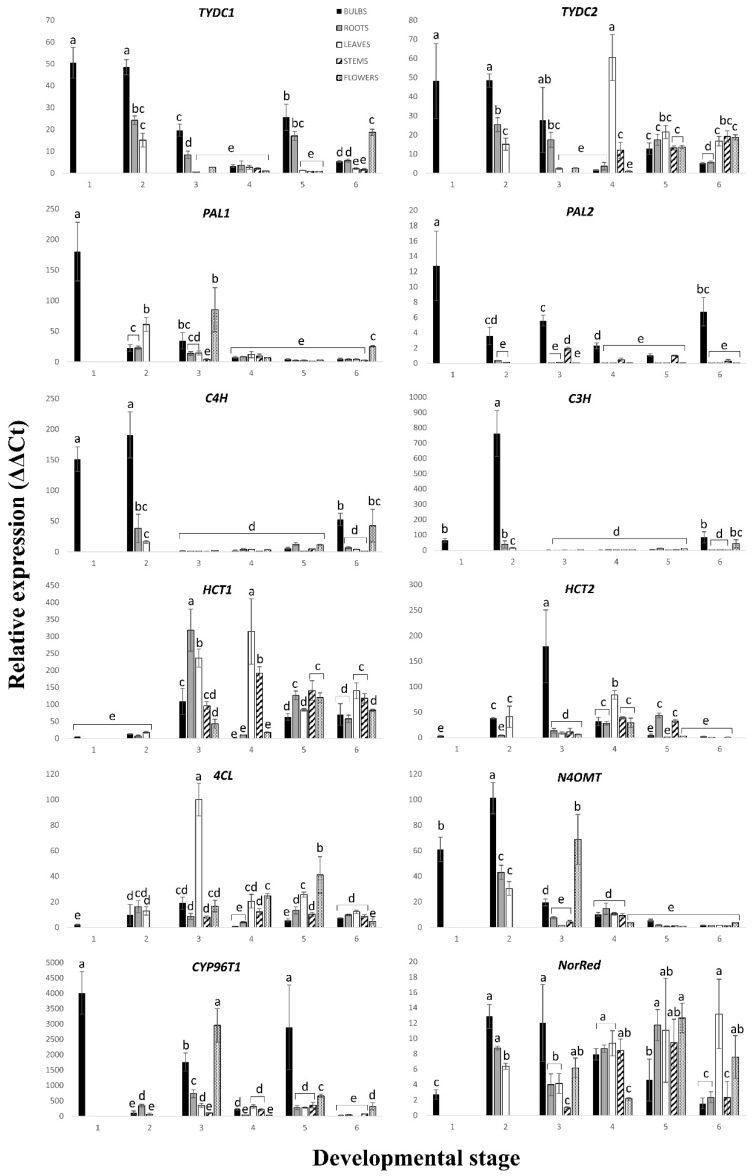
Figure 3.
Expression profiles of twelve genes involved in Amaryllidaceae alkaloid biosynthesis obtained by reverse transcription quantitative real-time PCR analysis of different parts (bulb, roots, leaves, stem and flowers) at six developmental stages of Narcissus papyraceus. The graphs are plotted using normalized ΔΔCT values scaled to the lowest value. Each graph shows the expression level of the gene indicated above the graph. The gene for histone was used as a reference gene. The gene expression fold change was calculated using the 2−ΔΔCT method [56]. The error bars represent the mean standard deviation of three independent replicates, which were calculated using the method stated in the CFX Connect Real-Time System manual [43]. Abbreviations are TYDC, tyrosine decarboxylase; PAL, phenylalanine ammonia lyase; C4H, cinnamate 4-hydroxylase; C3H, coumarate 3-hydroxylase; 4CL, 4-coumarate CoA ligase; HCT, hydroxycinnamoyl transferase; N4OMT, norbelladine 4′-O-methyltransferase; CYP96T1, cytochrome P450 monooxygenase 96T1 and NorRed, noroxomaritidine reductase. The developmental stages are detailed in Figure 2. The bars having the same letter present no significant differences (p < 0.05) according to the Tukey statistical test.