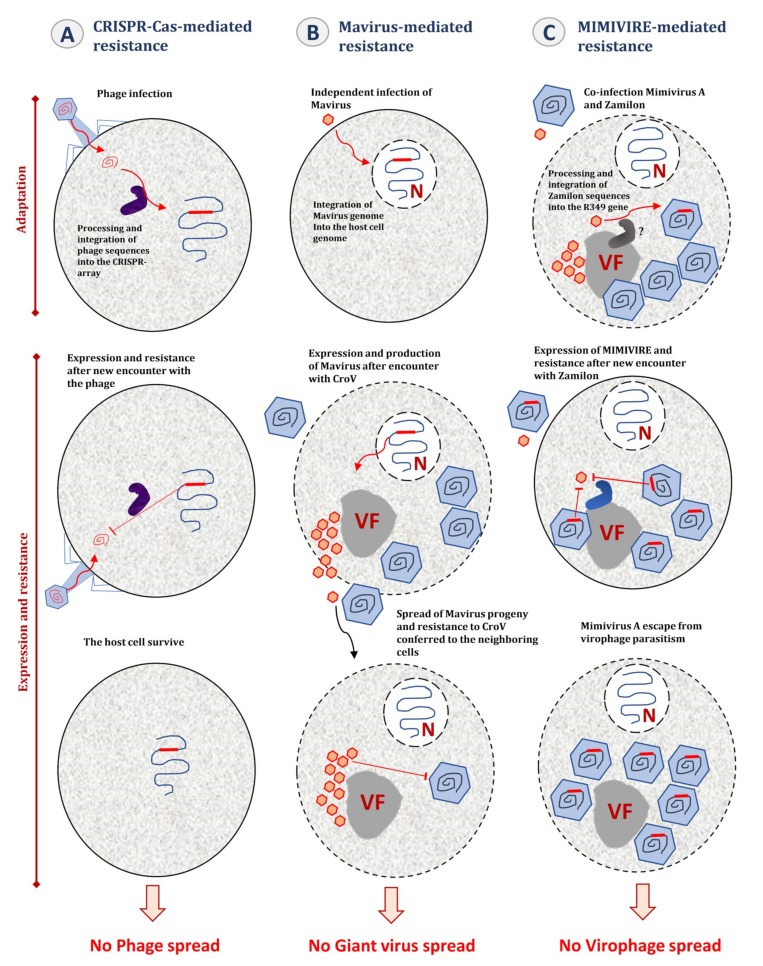
Figure 5.
Analogies between the CRISPR-Cas-, the Mavirus- immunity, and the MIMIVIRE-mediated immunity systems. All these mechanisms seem to implicate a record of genetic sequences and their integration into the genome of immunized organisms at their first step (adaptation). (A) For the CRISPR-Cas system, the host cell incorporates DNA pieces from the invading phage (proto-spacers) into the CRISPR arrays (spacers) (not represented here). This mechanism is mediated by the Cas machinery (in purple). (B) After an independent entry of Mavirus in the host cell, the virophage integrates its genome into the host genome and becomes latent. This mechanism seems to involve Mavirus-encoded retroviral integrase. (C) Mimiviruses from lineage A have integrated DNA sequences from Zamilon into their R349 genes. However, unlike the CRISPR-Cas and Mavirus systems, the integration mechanism of Zamilon DNA has not been characterized. The next step of the resistance mechanism is marked by the expression of the integrated genetic pieces. (A) For the CRISPR-Cas system, transcripts of the spacers are used as a guide for the Cas proteins to cut foreign DNA after new encounters with the phage. This phenomenon allows the host cell to survive and prevents the spread of the phage. (B) Infection of the host cell with Cafeteria roenbergensis virus (CroV) activates the expression and production of Mavirus. The host cell is lysed at the end of the CroV replication cycle but spread of the virophage progeny confers resistance to neighboring cells, allowing their survival and avoiding CroV spread. (C) The expression of the R349 gene containing Zamilon DNA and of the Cas-like proteins (in blue) is associated with the inhibition of Zamilon replication, conferring to lineage A mimiviruses a resistance to this virophage.