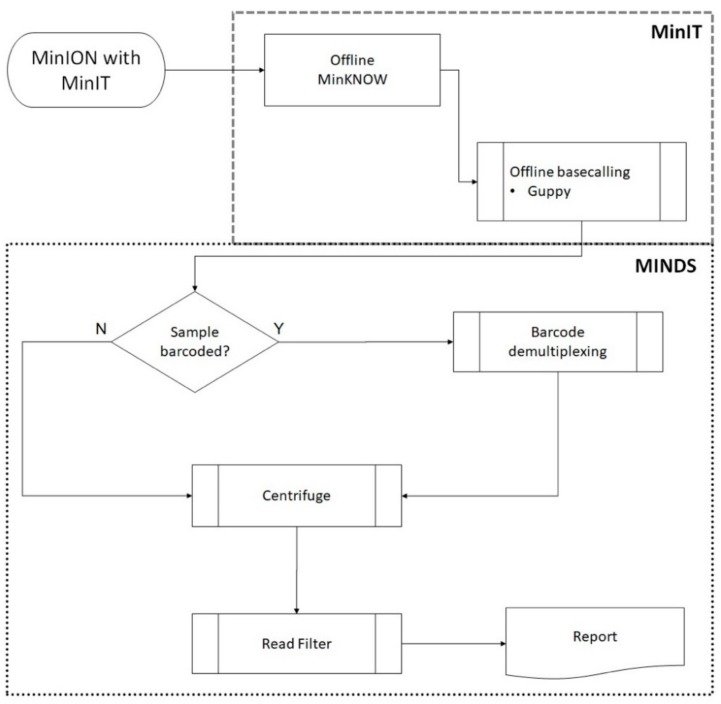
Figure 1.
Schematic demonstrating sequencing and bioinformatic workflow. Reads from the MinION were acquired with Oxford Nanopore Technologies’ (ONT’s) MinIT real-time base-calling device running MinKNOW and Guppy software. The downstream MINDS workflow processed the metagenomic sequence data read wirelessly from the MinIT to identify microorganisms present in the sample. Barcoded samples were first demultiplexed (if required) using ONT’s qcat software. Centrifuge (Johns Hopkins Center Computational Biology) mapped reads to taxonomic classifications. Background noise was filtered by removing species with only sporadic reads: having less than 0.1% of all total reads mapped and/or having less than 5 unique reads mapped.