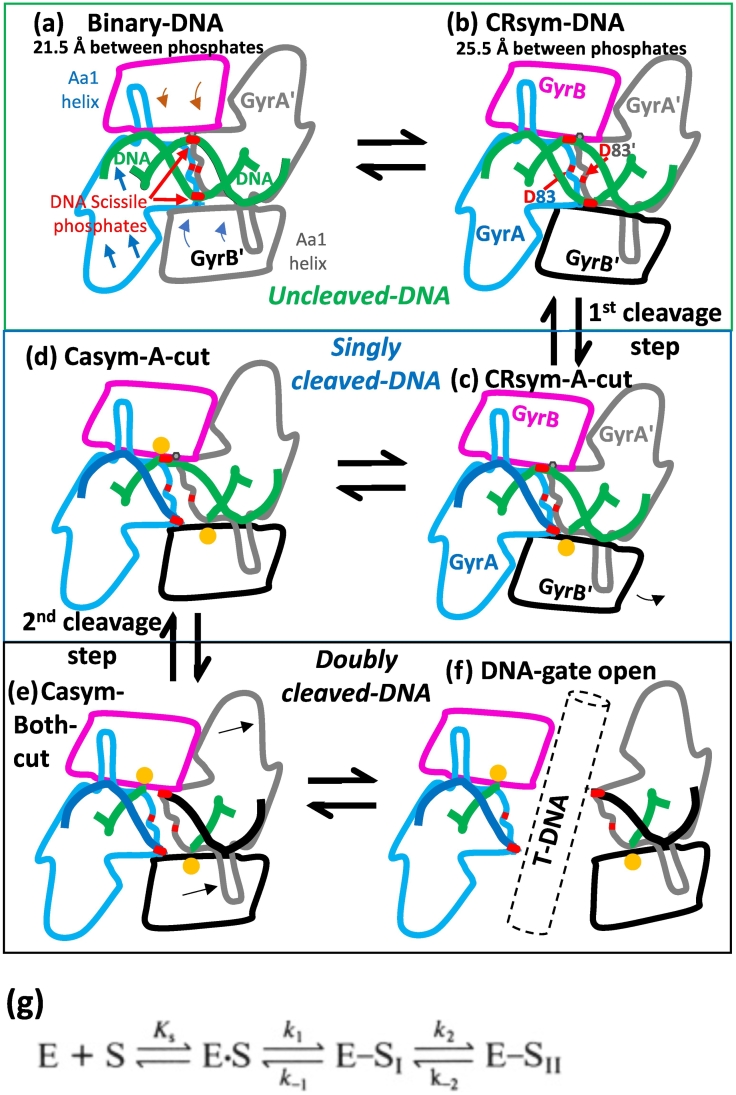
Fig. 8.
A simplified model for DNA cleavage by S. aureus DNA gyrase. (A) A G-DNA segment is shown in a binary complex, based on binary structure (6FQV). No metal is bound. The scissile phosphate (red) is outlined on the DNA backbone (green). D83 and D83’ are close to each other at the GyrA/GyrA’ dimer interface (red lines). (B) A T-DNA segment has been captured (not shown) and the domains at the DNA-gate start to move, stretching the substrate gate DNA. (C) A single metal (yellow sphere) is captured in the A-configuration, on one side. (D) The bottom strand has been cut, the metal is in the B-configuration, the gate is pushed further open and a metal is captured in the A-configuration on the other side. (E) Both DNA strands are now cleaved. Metals are in B-configurations. (F) The T-DNA is outlined passing through the DNA gate. (G) The above model is broadly consistent with the scheme published by Zechiedrich et al. [49]; E is the enzyme and S is the substrate DNA, which is either uncleaved (E.S—A,B), single cleaved (E-S1—C,D) or doubly cleaved (E-S11—E,F).