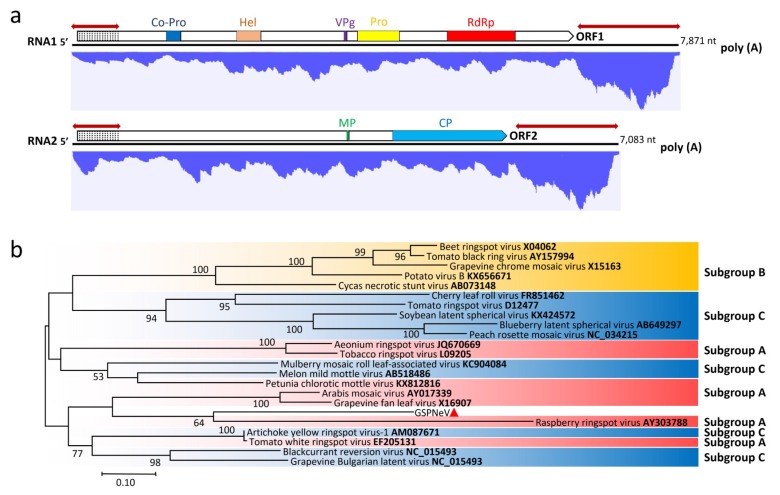
Figure 2.
(a) Genomic structure of green Sichuan pepper-nepovirus (GSPNeV) and distribution profile of transcriptomic reads from GSPNeV. The colorful blocks in putative proteins represent conserved amino acid domains or motifs. Co-Pro: Proteinase co-factor. Hel: Helicase. VPg: Viral protein linked to the genome. Pro: Proteinase. RdRp: RNA dependent RNA polymerase. Poly (A): A 3′ terminal poly (A) tail. Similar-sized double-headed red arrows indicate extensive regions of identical nucleotides between RNA1 and RNA2; dotted areas at 5′ end of coding region indicate extensive region of identical amino acids; (b) maximum-likelihood relationships [Jones-Taylor-Thornton (JTT) model] inferred from amino acid sequence alignments of the conserved CG-GDD region between Pro and RdRp domains of GSPNeV (marked with small red triangle) and representative nepoviruses divided into three subgroups (A, B, and C). Bootstrap support (as a percentage of 1000 replicates) is shown at the nodes for values greater than 50%. Accession numbers are supplied in taxa names.