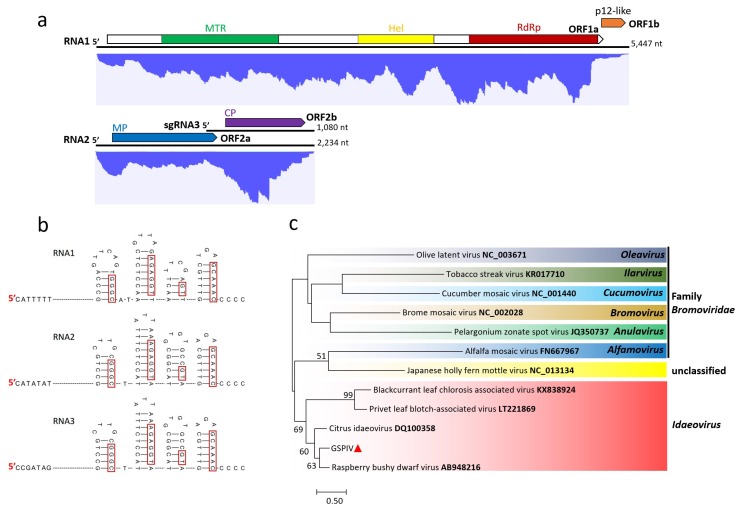
Figure 3.
(a) Genomic structure of green Sichuan pepper-idaeovirus (GSPIV) and distribution profile of transcriptomic reads from GSPIV. The colorful blocks in proteins represent conserved amino acid sequences, domains or motifs. MTR: Methyltransferase. Hel: Helicase. RdRp: RNA dependent RNA polymerase. p12-like: Putative p12-like protein. MP: Movement protein. CP: Coat protein. sgRNA3: Subgenomic RNA3; (b) first seven nucleotides of 5′ genomic terminus and, stem-loops and the last conserved five cytidines (C) of 3′ genomic terminus from RNA1, 2 and sgRNA3 are shown. The identical nucleotides at the stem shared among the RNAs are indicated with red boxes; (c) maximum-likelihood tree (JTT model) derived from entire CP amino acid sequence comparisons of GSPIV (marked with small red triangle), representative idaeoviruses, and related viruses. Bootstrap support values (as a percentage of 1000 replicates) are shown at the nodes if greater than 50%. The accession number is given in taxa names.