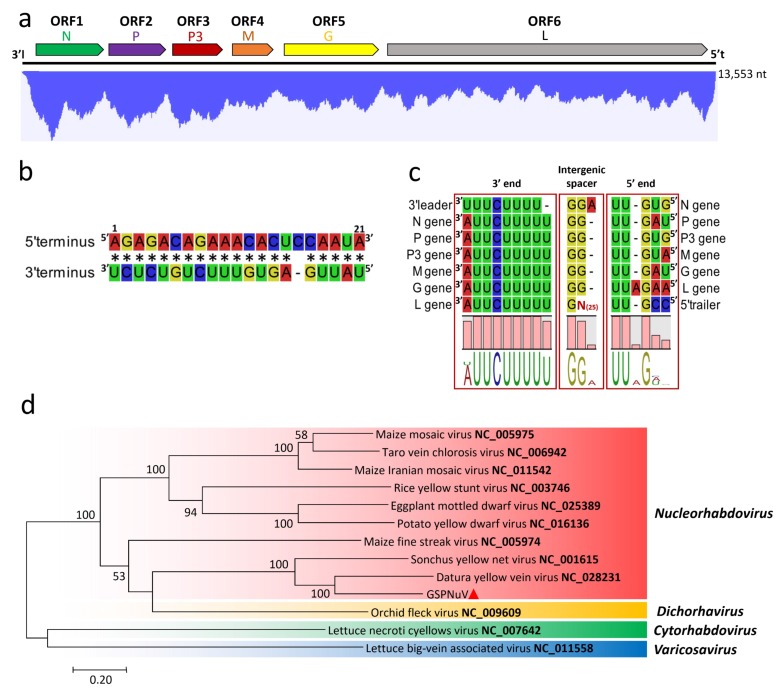
Figure 5.
(a) Genomic structure of green Sichuan pepper-nucleorhabdovirus (GSPNuV) and profile distribution of transcriptomic reads from GSPNuV. N, P, P3, M, G, and L are putative nucleocapsid protein, phosphoprotein, movement protein, matrix protein, glycoprotein, and polymerase, respectively. 3’l and 5’t are a leader sequence at 3′ genomic terminus and a trailer sequence at the 5′ terminus, respectively; (b) the complementary sequences of the 5′ and 3′ termini (asterisk indicates complementary bases); (c) conserved gene junction sequences interposed between two adjacent genes; (d) phylogenetic tree generated using the maximum-likelihood method (JTT model) and 1000 bootstrap replicates, based on alignment of amino acid sequences of the L genes from GSPNuV (marked with small red triangle) and related viruses. Bootstrap percent values greater than 50 are shown at the nodes. The viral accession number is marked in bold in taxa names.