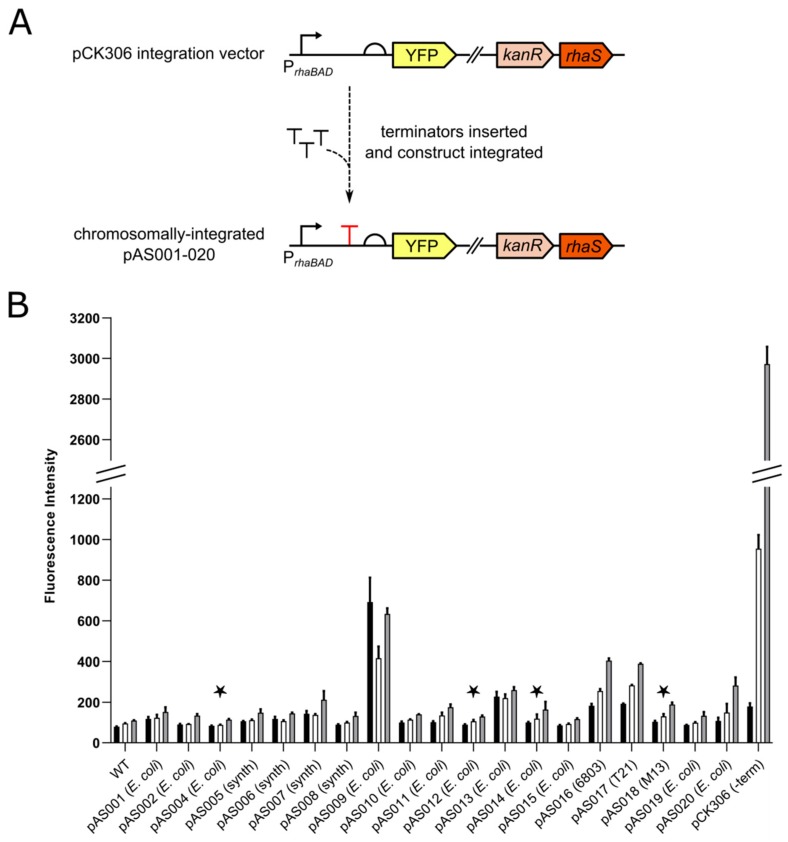
Figure 1.
Screening of terminators in Synechocystis. (A) Overview showing insertion of terminators into plasmid pCK306 between the rhaBAD promoter and the ribosome-binding site (RBS) of the yellow fluorescent protein (YFP)-encoding gene. The resulting terminator constructs pAS001, pAS002, pAS004–020 were integrated into the Synechocystis genome. (B) Synechocystis transformants containing the integrated terminator constructs were cultured in BG11 media supplemented with kanamycin and 0.6 mg/mL L-rhamnose in photoautotrophic conditions in constant light. Wild-type Synechocystis cells (WT) lacking YFP entirely and cells containing the YFP-encoding cassette of the parent plasmid pCK306 (no terminator between rhaBAD promoter and RBS of YFP-encoding gene) were used as controls. The fluorescence intensity (arbitrary units) of 10,000 cells from each culture was measured by flow cytometry at 0 h (immediately after the culture was inoculated), 48 h and 96 h (black, white and grey bars respectively). Error bars shown are the standard deviation of the mean for three independent biological replicates. Key for SBOL glyphs used in figure: right-angled arrow represents a promoter; T represents a terminator; semi-circle represents a ribosome-binding site (RBS); coloured blocks represent coding sequences or genes. Origin of each terminator in brackets after plasmid name: E. coli, synth (synthetic), 6803 (Synechocystis sp. PCC 6803), T21 (bacteriophage T21), M13 (bacteriophage M13). Stars above terminators refer to the four terminators that were used in the chromosomal insulation experiments (see Figure 2).