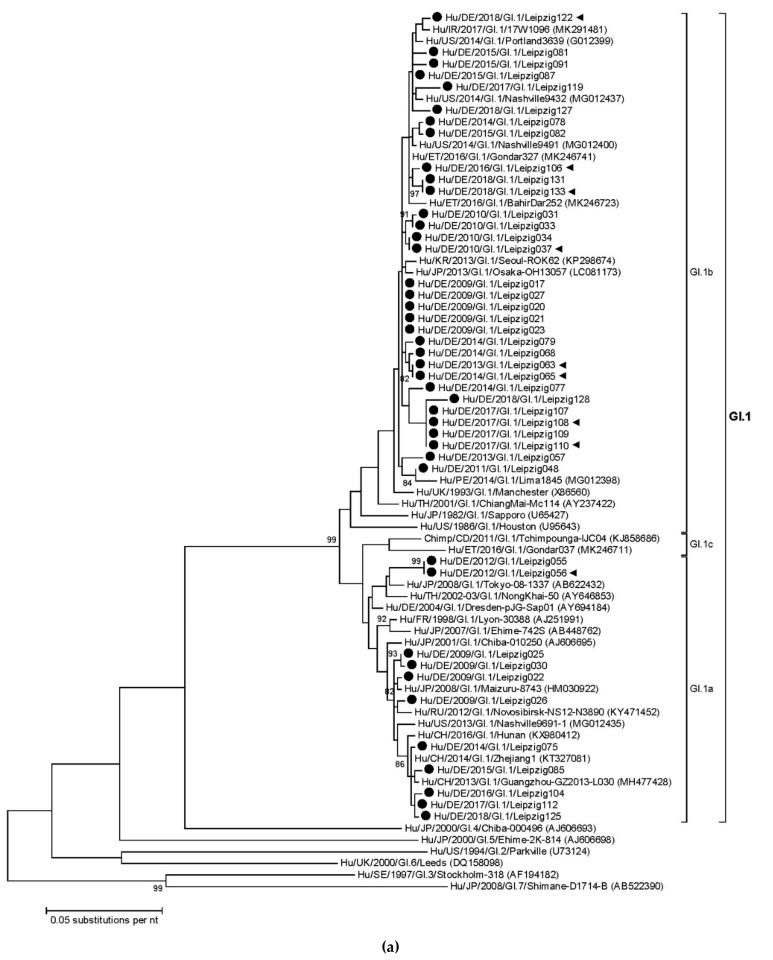
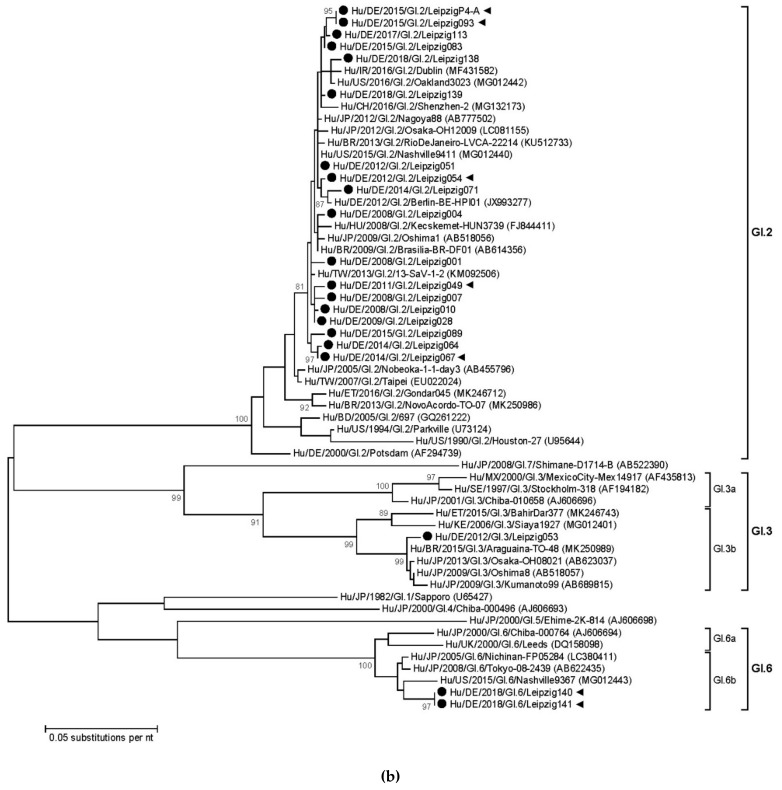
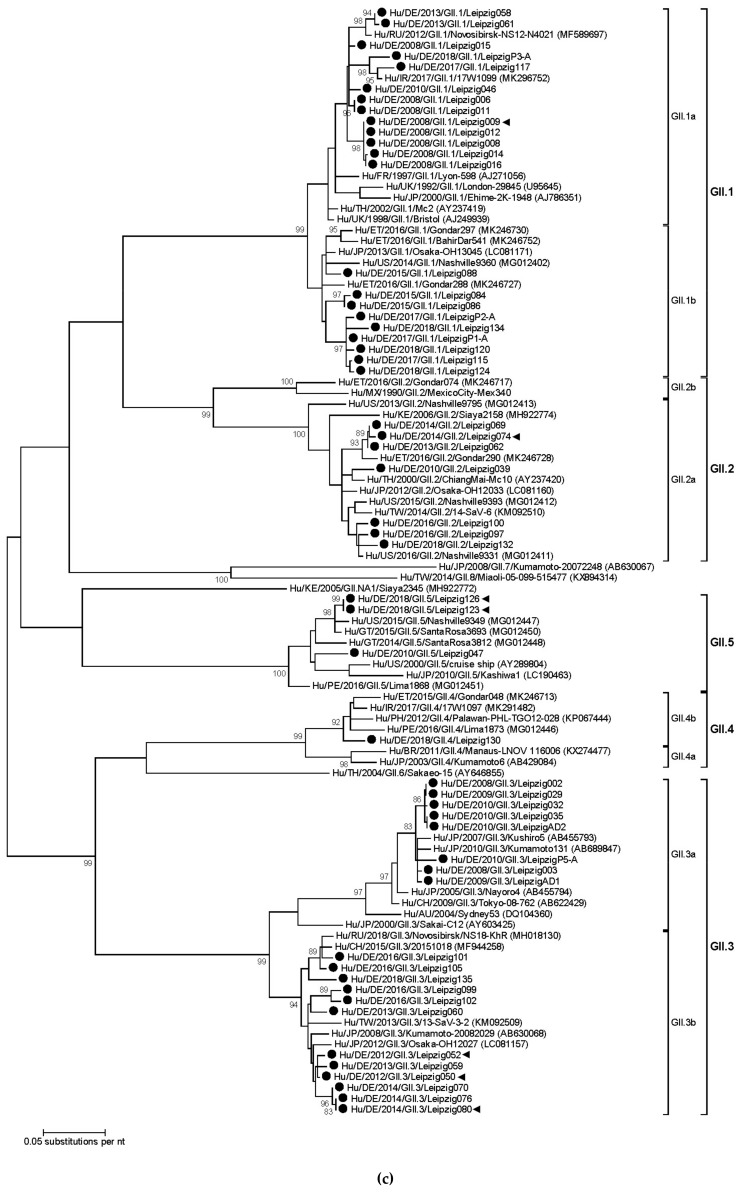
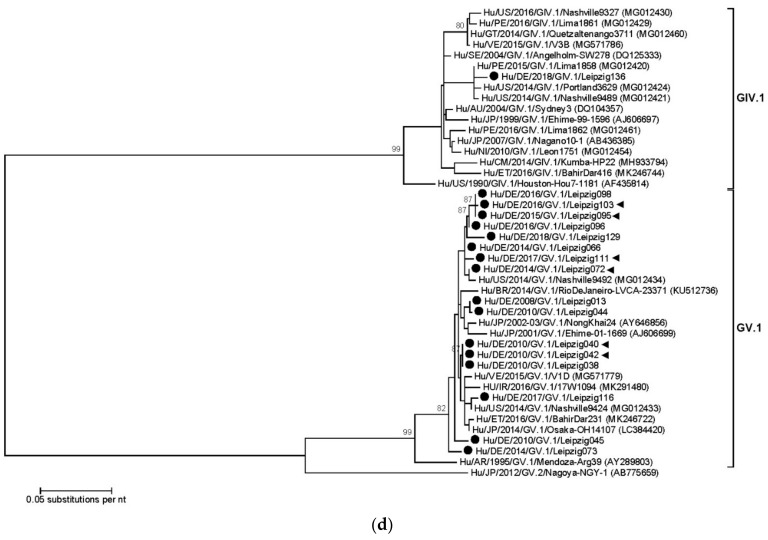
Figure 3.
Phylogenetic analysis of partial VP1 sequences (667−697 bp) at the nucleotide level. Separate trees are shown for (a) GI.1, (b) GI.2, GI.3, GI.4 and GI.6, (c) genogroup GII, and (d) genogroup GIV and GV sapoviruses. The trees were constructed using the Maximum Likelihood method implemented in MEGA5 software. Bootstraps values (1000 replicates) above 80% are shown. Sapovirus genotypes and intragenotypic lineages are indicated at the right. Strains of the present study are pointed out by black circles. Those detected in nosocomial sapovirus infections are additionally labelled with black triangles.