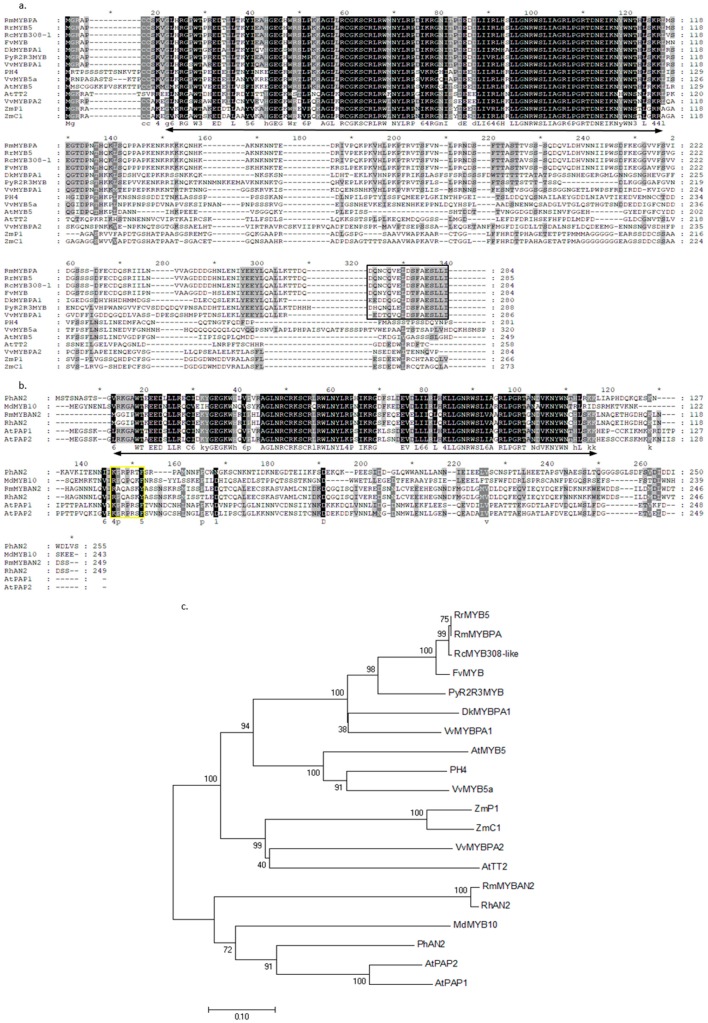
Figure 6.
Analysis of deduced amino acid sequences of two R2R3-MYB TFs. (a) Alignment of deduced amino acid sequences of the AN2 subgroup of MYB TFs, and (b) alignment of deduced amino acid sequences of the TT2 and PA specific-MYB regulators. ZmC1, ZmPl (maize), AtTT2, AtMYB5 (Arabidopsis thaliana), PH4 (Pharbitis hybrid), VvMYB5a, VvMYBPA1, VvMYBPA2 (Vitis vinifera), PyR2R3MYB (Prunus yedoensis), DkMYBPA1 (Diospyros kaki), RrMYB5 (Rosa rugusa), RhMYB308-like (Rosa chinensis), and FvMYB (Fragaria vesca). The R2R3MYB domains are indicated by black arrow, yellow box indicates a C-terminal conserved motif [K/R]Px3[K/T][F/Y] in AN2 subgroup of R2R3MYB. Black box indicates a C-terminal conserved motif in the PA-specific MYB regulators. (c) A neighbor-joining phylogenetic tree of plant R2R3MYB sequences. The scale bar represents 0.1 substitutions per site and the numbers next to the nodes are bootstrap values from 1000 replicates. The deduced amino acid sequences were retrieved from the DDBJ/EMBL/GeneBank databases, and the accession numbers are as follows AtPAP1(NP_176057.1), AtPAP2(NP_176813.1), RhAN2(BAM36703.1), PhAN2(AAF66727.1), MdMYB10(AAF66727.1), and ZmC1(sp|P10290.1).