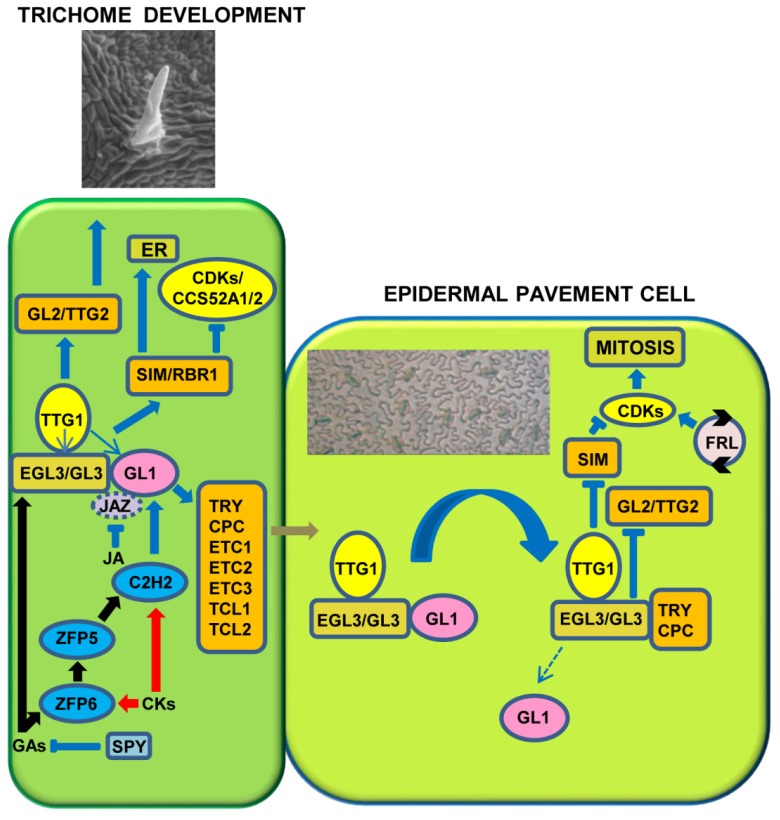
Figure 1.
A simplified model for the acquisition of the competence of epidermal pavement cells to become trichomes in the model species Arabidopsis thaliana. In epidermal pavement cells, GLABRA3 (GL3) acts together with GLABRA1 (GL1) and TRANSPARENT TESTA GLABRA1 (TTG1), creating a trimeric MYB/bHLH/WD (MBW) activator complex. TTG1 works upstream of GL3 and GL1, activating their expression (thin blue arrows). Gibberellins (GAs), cytokinins (CKs), and jasmonic acid (JA) contribute positively to the regulation of trichome development. GAs activate the transcription of the ZINC FINGER PROTEIN 6 (ZFP6) gene, a member of the large C2H2 regulatory gene family (black arrow). ZFP6 induces the expression of ZFP5, and then ZFP5 promotes GLABROUS INFLORESCENCE STEMS (GIS) (C2H2), GIS2, and ZFP8 expression (black arrows). At the same time, CKs promote ZP6, ZFP8, and GIS2 expression (red arrows). C2H2 members (i.e., GIS2 and ZFP8) increase the transcription of GL1 (blue arrow). The transcription levels of GL3, TTG1, and TRY are regulated by GAs (in the figure a single black arrow indicates the activation of the MBW complex). The SPINDLY (SPY) gene inhibits the GA signal (blue inhibitory line). JA regulates the formation of trichomes, favoring the degradation of proteins of the Jasmonate ZIM-domain (JAZ) (blue inhibitory line and blue dotted line); therefore, this hormone inhibits the interaction between JAZ with GL1 and EGL3/GL3 transcription factors (TFs). The MBW complex stimulates the development of trichomes, switching the transcription of several targets (represented by orange boxes): (i) SIAMESE (SIM) and RETINOBLASTOMA RELATED1 (RBR1) as key genes in cell cycle regulation (blue arrow); (ii) GL2 and TTG2 as positive regulators (blue arrow); and (iii) R3-MYB TFs (CAPRICE (CPC), TRYPTICON (TRY), ENHANCER OF TRY AND CPC1 (ETC1), ETC2, ETC3, TRICHOMELESS1 (TCL1) and TCL2) as repressors of trichome initiation and growth (blue arrow). SIM and RBR1 promote the trichome initiation from epidermal pavement cells through the down-regulation of some cyclin-dependent kinases (CDKs) and the CELL CYCLE SWITCH 52A1 (CCS52A1) and CCS52A2 genes, respectively (blue inhibitory line). Therefore, the conversion of a normal mitosis into the endoreduplication cycle (ER) is promoted (blue arrow). The R3-MYB inhibitors are capable of movement in adjacent cells (grey arrow) and replace GL1 in the MBW complex (bent and dotted blue arrows) to form a repressor complex, which cannot promote GL2 and TTG2 expression (blue inhibitory line), thereby inhibiting trichome fate. The repressor complex also loses ability to activate the target SIM gene (blue inhibitory line). A complex feedback regulatory loop (FRL, pink circle with black arrowheads), including, among other inhibitors of CDKs such as KIP-RELATED PROTEINS (KRPs) and SIM, RBR1 and E2F, TFs activate (blue arrow) some CDKs allowing cells entry into the mitotic cycle (blue arrow). TFs: transcription factors.