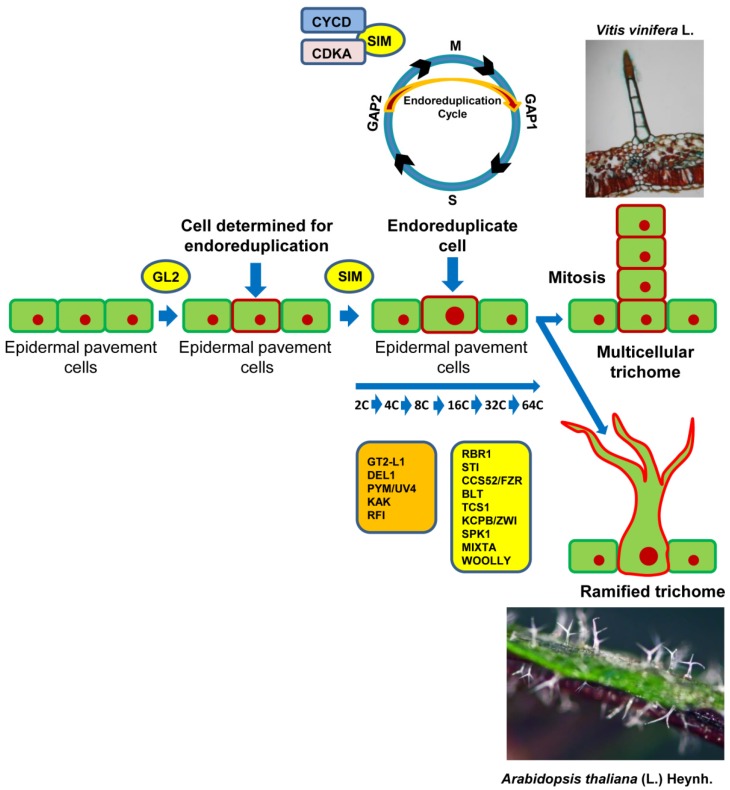
Figure 2.
A simplified model for the differentiation of multicellular and ramified trichomes. An epidermal pavement cell activated by GLABRA2 (GL2) becomes determined for trichome initiation (red cell wall). The SIAMESE (SIM) gene promotes the endoreduplication cycle (see also Figure 1). SIM interacts with D-type cyclin-CYCLIN-DEPENDENT KINASE A (CYCD-CDKA) complexes, which normally operate at the G1/S and G2/M transitions, to suppress entry into phase M. The nuclear DNA content increases from 2C to 64C. The endoreduplicate cell can follow two fates, also in relation to the species: entering the mitosis process, originating a multicellular trichome as in the grape (Vitis vinifera L.), or promoting the development of branched trichomes, as in Arabidopsis thaliana (L.) Heynh. In the orange box are indicated some negative regulators involved in the development of complex trichomes: GT2-LIKE1 trihelix (GT2-L1), DP-E2F-Like1 (DEL1), KAKTUS (KAK), POLYCHOME//UV-INSENSITIVE4 (PYM/UVI4), and RASTAFARI (RFI). By contrast, in the yellow box are indicated some positive regulators: RETINOBLASTOMA RELATED1 (RBR1), STICHEL (STI), CELL CYCLE SWITCH 52A2/FIZZY-RELATED1 (CCS52A2)/FZR1, CCS52A1/FZR2, BRACHLESS TRICHOME (BLT), TRICHOME CELL SHAPE 1 (TCS1), KINESIN-LIKE CALMODULIN-BINDING PROTEIN/ZWICHEL (KCBP/ZWI), BRACHLESS TRICHOME (BLT), SPIKE1 (SPK1), MIXTA and WOOLLY (WO). (See the text for the specific activity of each gene on cell cycle regulation). G1: Gap 1; G2: Gap2; M: mitosis, S: DNA synthesis.