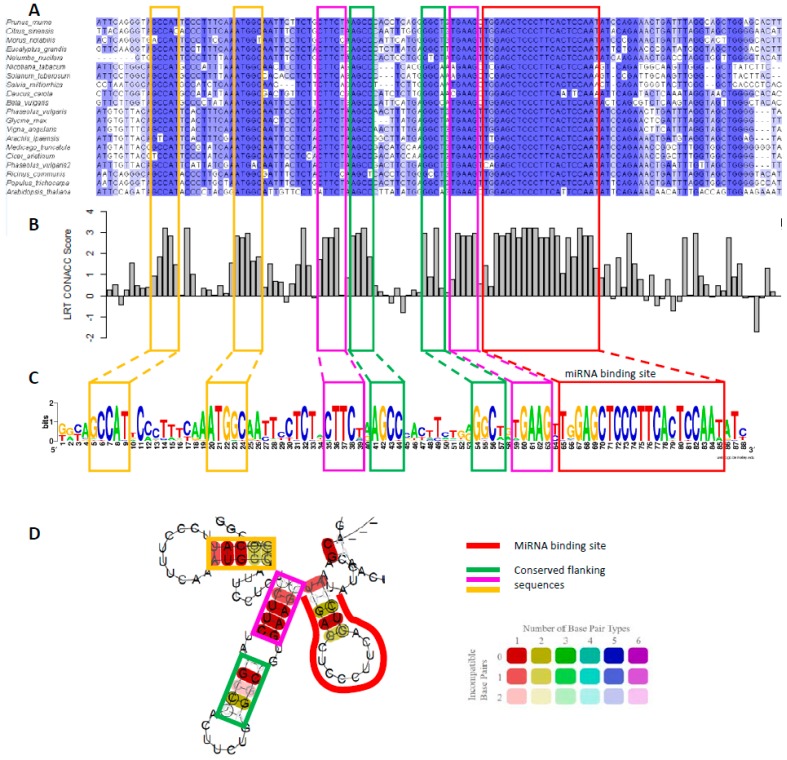
Figure 3.
(A) Multiple alignment of MYB33 homologues from different plants species. The binding site is boxed in red, and the conserved flanking sequences in yellow, pink, and green throughout the Figure. (B) phyloP score of the multiple sequence alignment of MYB33 sequences. A positive score denotes evolutionary conservation, whereas, a negative score denotes acceleration [54]. A likelihood ratio test (LRT) was used as the method to detect non-neutral substitution rates. Scores were generated using rPHAST [55]. (C) Sequence logo of the binding site and conserved flanking sequences generated using WebLogo [56]. (D) RNA secondary structure prediction of the consensus sequence from the multiple alignment in A. generated using RNAalifold [57] at 22 °C and default parameters. Colours represent the number of base pairs types (i.e., AU, UA, CG, GC, UG, GU), and hue the number of non-conserved nucleotides at that position.