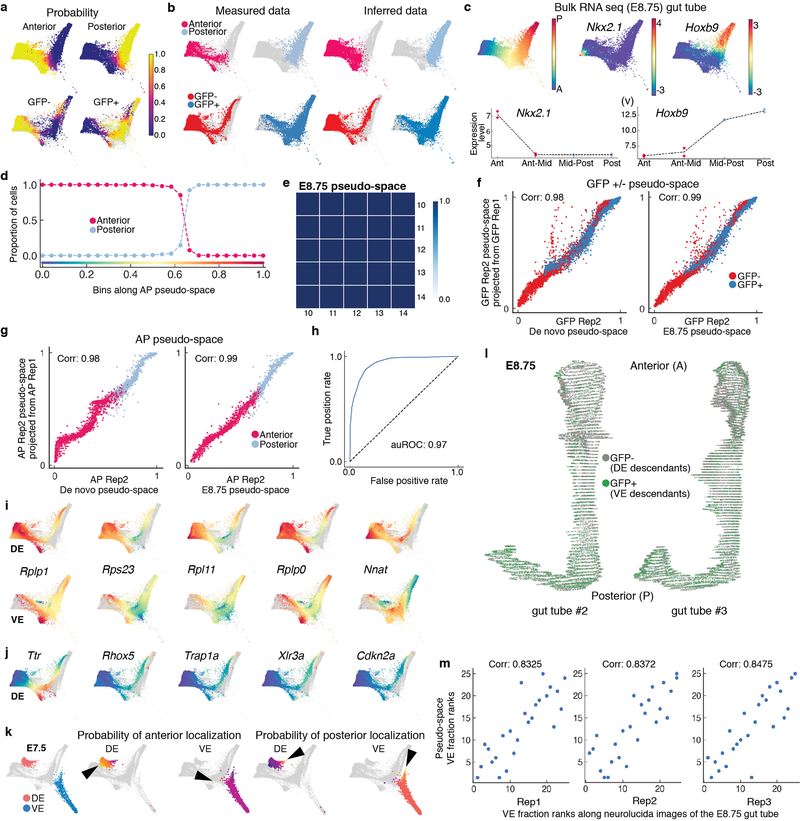
Extended Data Figure 8: Characterization of E8.75 gut tube anterior-posterior pseudo-space.
a, Force-directed layout as in Fig 5. (i): Plots showing the probabilities of anterior-posterior positioning for the Afp-GFP-positive/Afp-GFP-negative cells inferred using the manifold classifier trained on anterior-posterior cell. (ii): Plots showing the probabilities of GFP-positive/GFP-negative status for the cells from the anterior-posterior compartment inferred using the manifold classifier trained on GFP-positive/GFP-negative cells. b, (i): Anterior and posterior cells labeled by measured data (left). Anterior and posterior positions of Afp-GFP-positive/AFP-GFP-negative cells inferred data (right) using probabilities in (a-i). (ii): GFP-positive/GFP-negative cells labeled by measured data (left). GFP-positive/GFP-negative status of the anterior-posterior compartment cells inferred using probabilities in (a-ii). c, (i): Plot showing the first diffusion component of the E8.75 cells. (ii-iii): Plots showing the expression of anterior marker Nkx2.1 and posterior marker Hoxb9 in E8.75 cells. (iv-v): Bulk RNA-seq expression of Nkx2–1 and Hoxb9 in quadrants of the gut tube along the AP axis compares with A-P single cell expression patterns. d, Plot showing the proportion of anterior and posterior cells in bins along the AP pseudo-space axis. e, Receiver operating curve for classification of E7.5 VE and DE cells (4378 cells). f, Plots showing the expression patterns of genes that are best predictive of the DE class in the VE-DE classifier (top - DE; bottom -VE). g, Plots showing the expression patterns of genes in the DE best predictive of VE class in the VE-DE classifier. h, Force directed layouts following Harmony of E7.5 and E8.75 VE and DE cells with E7.5 cells highlighted in red (DE) and blue (VE) (left). E7.5 VE and DE cells colored by the branch probability of anterior localization (middle) and posterior localization (right). Black arrowheads indicate early emergence of AP spatial patterning at E7.5, with E7.5 DE cells predominantly destined towards anterior, and VE cells predominantly destined towards posterior. i, 3D renderings of gut tube depicting all endoderm cells along AP axis. Nuclei of VE and DE cells are labeled in green and grey, respectively. j, Plots comparing the ranks of proportion of GFP-positive cells along AP positioning in the Afp-GFP embryo-derived Neurolucida gut tube replicates (x-axis), and the ranks of VE cell proportions in bins along the AP pseudo-space axis (y-axis), the AP axis was partitioned into 20 bins, each dot representing the fraction of VE cells in that bin. k, Heatmap showing Pearson correlations between AP pseudo-space orderings determined using a varying number of diffusion components highlighting the robustness of the ordering. l, Plots comparing the AP pseudo-space ordering of GFP-positive/GFP-negative cells (replicate 2: 13335 cells) generated de novo using only the replicate 2 cells (x-axis, left) with the projected ordering from replicate 1 (8143 cells) (y-axis). Right panel shows a similar comparison with the pseudo-space ordering determined using cells of both the replicates on the x-axis. m, Same as l, for replicates of anterior-posterior cells (Replicate 1: 1821 cells, replicate 2: 1691 cells). Plots show the Pearson correlation.