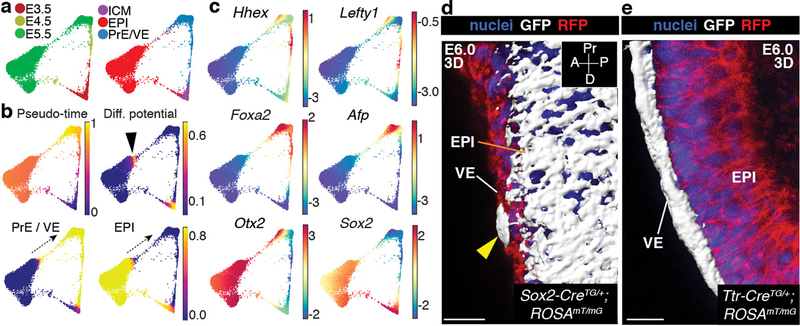
Figure 2: Differentiation of epiblast into endoderm before gastrulation.
Results from Harmony applied to all replicates of E3.5-E5.5. a, Force directed layouts depicting relationship between EPI and PrE/VE lineages. Cells colored by time-point (left) and cell type labels (right). b, Palantir pseudo-time, differentiation potential (DP) and branch probabilities (BP) of EPI and PrE/VE cell lineages. Black arrowhead and dotted arrows denote EPI cells with high DP, representing a trans-differentiation to endoderm. c, Gene expression of AVE (Hhex, Lefty1), VE (Foxa2, Afp), VE and EPI (Otx2, Sox2) markers. Cells colored based on post-MAGIC50 gene expression. d–e, 3D surface renderings of mGFP-expressing cells in E6.0 Sox2-CreTG/+;ROSA26mT/mG (d) and Ttr-CreTG/+;ROSA26mT/mG embryos (e). Nuclei stained with Hoechst, membranes labeled with RFP. Results validated in >3 independent experiments. ExE, extra-embryonic ectoderm. Scale bars: 10μm A, anterior; D, distal; P, posterior; Pr, proximal.