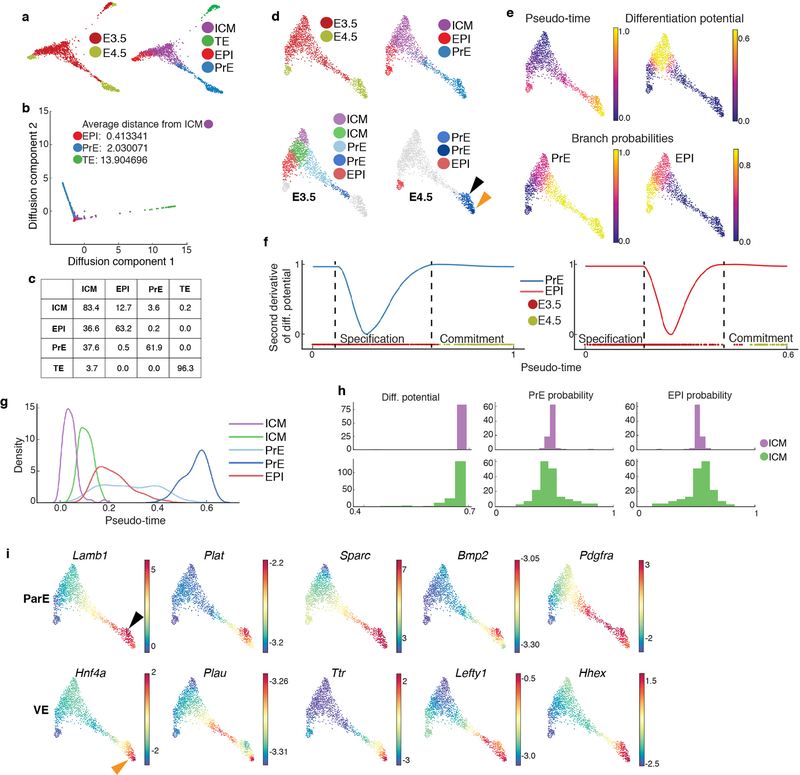
Extended Data Figure 4: Lineage decisions in the mammalian blastocyst.
Results from pooling cells of two replicates of E3.5 and E4.5 followed by Harmony augmentation. a, Force directed layout of E3.5 and E4.5 cells depicting relationship between three blastocyst lineages. Cells colored by time-point or annotated cell types. b, Plot showing projection of E3.5 and E4.5 cells along first two diffusion components. Distances between lineages were computed using multi-scale distances. c, Table showing the connectivity between different compartments in a kNN graph of E3.5 cells. Each row represents the fraction of outgoing edges from cells of the respective compartment connecting to cells in the compartments specified in the columns. d, Force directed layout of E3.5 and E4.5, following removal of TE cells, showing relationships between ICM, EPI and PrE. Cells are colored by time-point or Phenograph14 clusters. e, Palantir10,11 determined pseudo-time ordering, differentiation potential (DP) and branch probabilities (BP) of PrE and EPI cell lineages. f, Plots showing the second derivative of PrE and EPI differentiation potential along pseudo-time suggesting that changes in differentiation potential, and hence lineage commitment in both lineages occur at E3.5. Points of highest changes along pseudo-time represent inferred lineage specification and commitment. g, Distribution of E3.5 lineage cells along pseudo-time, each distribution represents cells from one Phenograph cluster. h, Histograms showing the distribution of differentiation potential (left), PrE fate probability (middle) and EPI fate probability (right) in the E3.5 ICM clusters. i, Gene expression patterns of parietal (ParE) and visceral endoderm (VE) markers. Each cell is colored based on its MAGIC15,50 imputed expression level for the indicated gene. Black and orange arrowheads mark presumptive ParE and VE lineages, respectively.